Figure 6. Chimp ZFP568 has reduced binding affinity to its own Igf2-P0.
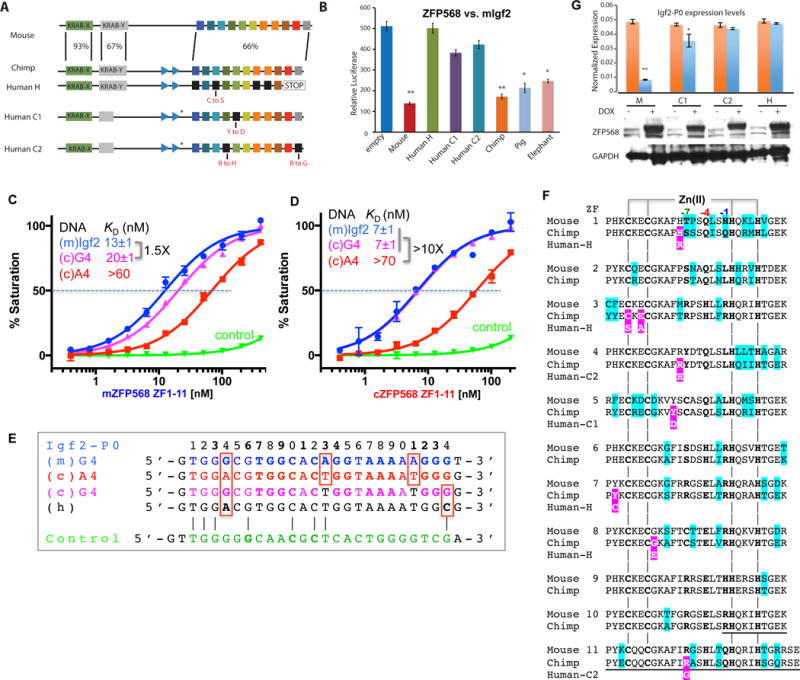
(A) Zfp568 ortholog in chimpanzee compared with mouse with percent conservation within each domain indicated, including the mutation N-terminal to ZF1 in the linker region (marked with a *).
(B) Luciferase assay to test mouse, human alleles H, C1, C2, chimp, pig and elephant ZFP568 repression activity against the mouse Igf2-P0 binding site cloned upstream of an SV40 promoter. Data in panels B and G are shown as mean±SD, t-test, *p<0.05, **p<0.01, n=3.
(C) The A4-to-G4 change at position 4 shows enhanced DNA binding by mZFP568.
(D) The A4-to-G4 change at position 4 recuperates the DNA binding by chimp ZFP568 to the same magnitude as that of mouse Igf2-P0 sequence.
(E) Chimp Igf2-P0 (A4 sequence) deviates from mouse Igf2-P0 at three locations including an Ade instead of Gua at position 4.
(F) Pairwise sequence alignment between mouse and chimp ZFP568 orthologs. The sequence variations are highlighted in cyan. The deviations of human alleles from the chimp sequence are highlighted with white letters against magenta. The underlined sequences in ZF10 and ZF11 are deleted in human allele H.
(G) Zfp568KO/KO ESCs were “rescued” by infection with DOX inducible lentiviral constructs expressing mouse ZFP568 (M) or human ZNF568 proteins encoded by the H, C1, or C2 alleles. ZFP568 Western blots (below) were performed to demonstrate similar expression levels of the rescue constructs. The relative levels of Igf2-P0 were determined by RT-qPCR.
