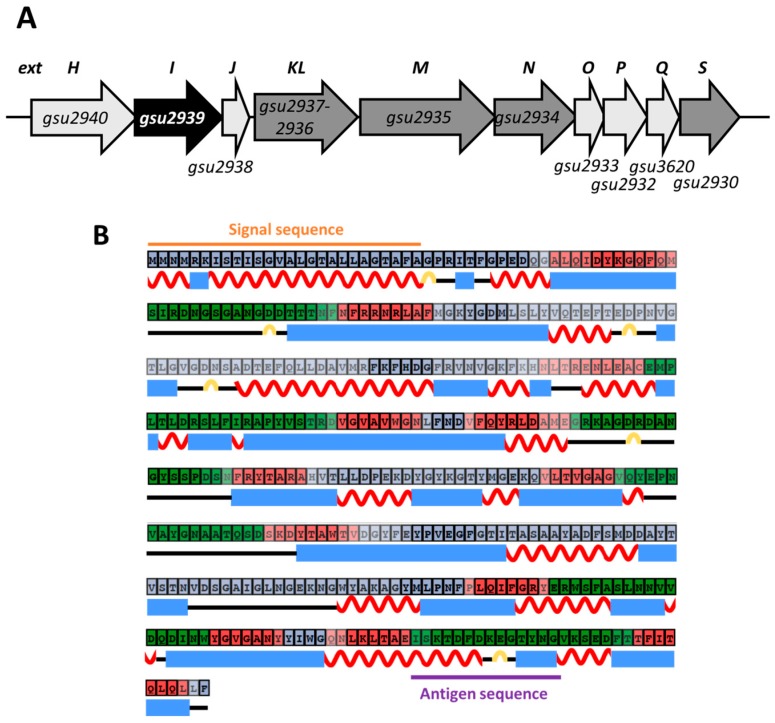
Figure 1.
Bioinformatics analysis of ExtI. (A) The extHIJKLMNOPQS gene cluster in G. sulfurreducens. The black arrow indicates the gene for a porin-like protein, extI (gsu2939), and the dark grey arrows indicate the genes for putative Cytc proteins, extKL (gsu2937-2936), extM (gsu2935), extN (gsu2934), and extS (gsu2930). The light gray arrows indicate the genes for a rhodanese-like protein, extH (gsu2940); a small protein, extJ (gsu2938); a Rieske iron-sulfur protein, extO (gsu2933); a cytochrome b, extP (gsu2932); and a membrane protein, extQ (gsu3620). (B) Prediction of the secondary structure of the ExtI protein. The amino-acid sequence of ExtI was computationally analyzed, and the secondary structure and the signal region were predicted. The N-terminal signal sequence (1–26 a.a.) predicted by SignalP 4.1 and the transmembrane α-helix region in the signal sequence (7–26 a.a.) predicted by TMHMM are depicted. Amino-acids in red, green, and gray boxes are predicted to be transmembrane, extracellular side, and periplasmic side, respectively. Red wavy lines, blue squares, and yellow lines represent α-helices, β-strands, and turns, respectively. The purple line shows the peptide sequence of the antigen used for preparation of the anti-ExtI antibody.