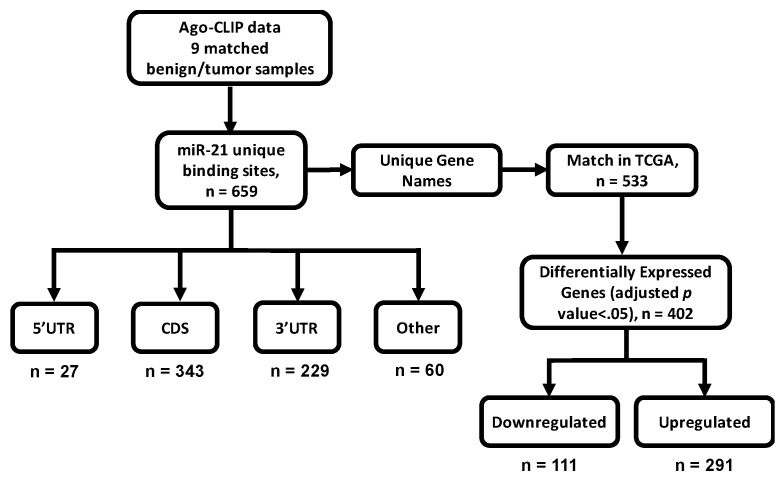
Figure 1.
Workflow for the selection and analysis of microRNA-21 (miR-21) targets. Unique miR-21 targets (n = 659) were identified using high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation of Argonaute (Ago-CLIP) data generated from matching benign liver and tumor tissues isolated from nine hepatocellular carcinoma (HCC) patients (GSE97061) [21]. Targets were sorted based on annotation of target loci: 5′ untranslated region [5′UTR], coding sequence [CDS], 3′ untranslated region [3′UTR], or other. Other groups included were transposable elements, introns, etc. Unique targets were compared to RNA-seq data generated by The Cancer Genome Atlas (TCGA) after log2 transformation and differential expression analysis between benign tissue and tumor tissue using the limma package in R [22]. Significance for differentially expressed genes was defined as having an adjusted p-value < 0.05 (n = 402).