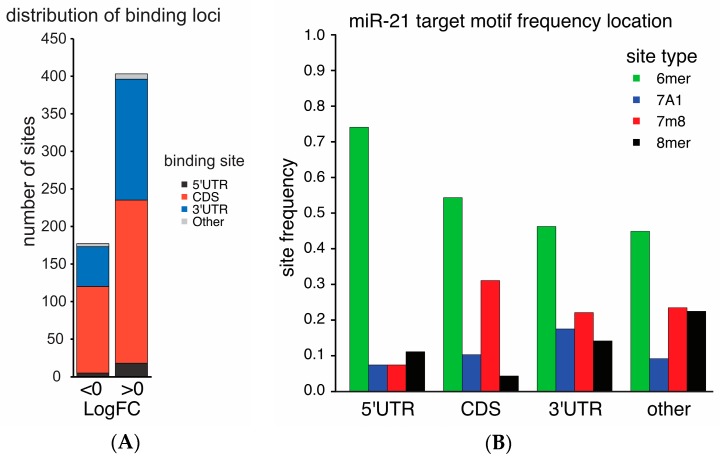
Figure 2.
Binding sites for miR-21 are most commonly in the CDS and 3′UTR of mRNA with variation in binding motif frequency. miR-21 binding sites identified by Ago-CLIP were classified by the sign of their log-fold enrichment in HCC and by the location of miR-21 binding on the target transcript: 5′ untranslated region, CDS, 3′UTR or other (A). The binding sites of miR-21 were classified by the frequency of the target sequence motif (6mer, 7A1, 7m8, 8mer) at each annotated location (B). The distribution of sequence motifs varied between the CDS and the 3′UTR (p = 2.1 × 10−6).