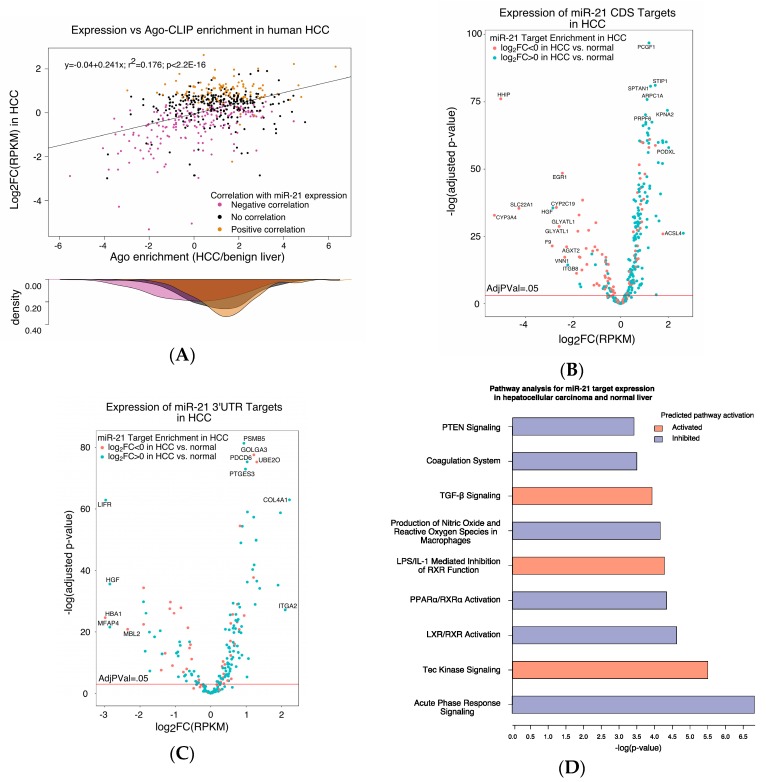
Figure 4.
The expression data of the miR-21 interactome in tumor (n = 377) and matched normal tissue (n = 59) in patients with liver cancer were extracted from TCGA and compared to the tumor enrichment score determined by Ago-CLIP (A). Log2-fold enrichment of miR-21 targets by Ago-CLIP in HCC positively correlated with log2-fold expression change in tumor (p < 2.2 × 10−16). Genes whose expression in tumors negatively correlated with miR-21 expression were less enriched in HCC by Ago-CLIP than those with positive correlation (p < 2.2 × 10−16). Among miR-21 CDS targets, 64 genes were significantly downregulated and 173 genes were significantly upregulated (B). Among miR-21 3′UTR targets, 48 genes were significantly downregulated and 173 genes were significantly upregulated (C). Expression data for miR-21 targets in HCC were subjected to Ingenuity Pathway Analysis (IPA). Pathways predicted to be activated or inhibited by the miR-21 targetome are shown (D). Pathway significance was defined as having an adjusted p-value < 0.0005.