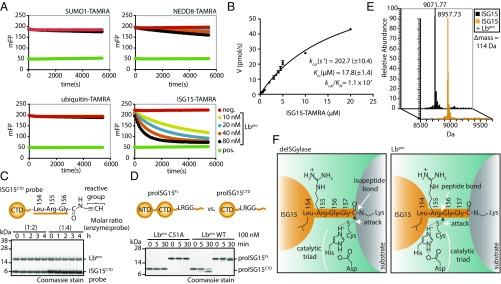
Fig. 1.
Lbpro substrate specificity and cleavage. (A) Specificity analysis of Lbpro against ubiquitin and ubiquitin-like (SUMO1, NEDD8, ISG15) TAMRA substrates (Fig. S1). (B) Michaelis–Menten kinetics of Lbpro as measured by ISG15-TAMRA cleavage. Error bars represent SD from the mean. (C) Suicide ISG15 probe assays were performed with Lbpro and WT ISG15CTD probe. (D) Comparison of full-length proISG15 (amino acids 1–165) and proISG15CTD (amino acids 79–165) cleavage by Lbpro. (E) Electrospray ionization MS of untreated and Lbpro-treated mature ISG15CTD (amino acids 79–157). The difference in mass corresponds to loss of the Gly-Gly peptide (114 Da). (F) Schematic of ISG15 cleavage by a canonical deISGylase (Left) and Lbpro (Right). deISGylases cleave isopeptide bonds, while Lbpro cleaves the peptide bond C-terminal to Arg155, releasing a GlyGly dipeptide. All assays were performed in triplicate. NTD, N-terminal ubiquitin-like domain.