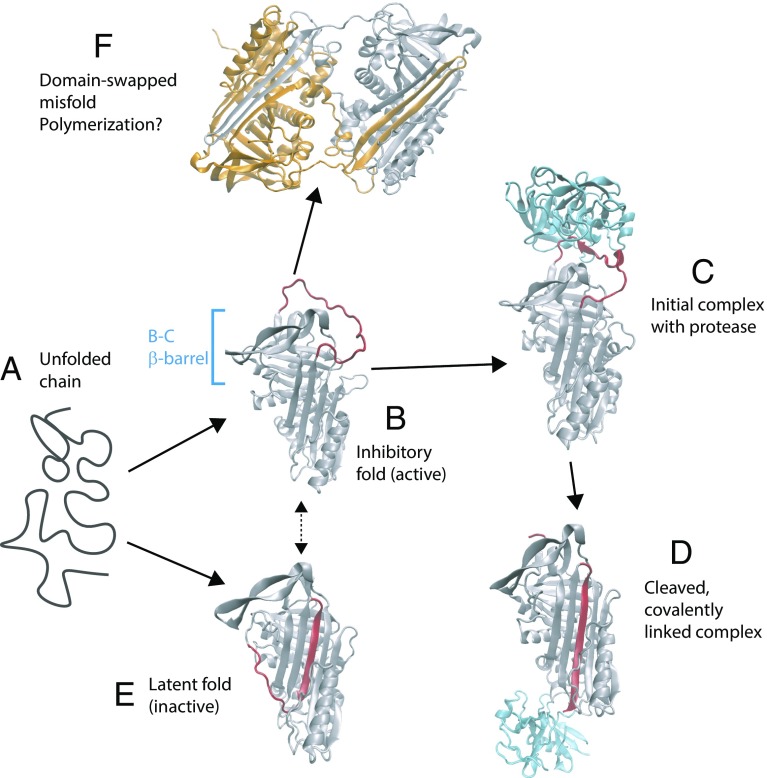
Fig. 1.
Stable states populated by the serpin α1-antitrypsin. The unfolded protein (a) initially folds to an active, inhibitory conformation (b) in which the RCL (red) is exposed [Protein Data Bank (PDB) ID code 1QLP (19)]. (c) Protease (in this case, trypsin, shown in blue) binds to the RCL [PDB ID code 1OPH (20)]. (d) Loop is cleaved by the protease, resulting in the enzyme being covalently linked to the serpin and insertion of the RCL sequence into a β-sheet [PDB ID code 1EZX (21)]. (e) Serpin can also adopt a latent fold, similar to the cleaved structure, which is more thermodynamically stable than the initially formed inhibitory fold [PDB ID code 1DVN (22)]. (f) Domain-swapped dimer structure [PDB ID code 2ZNH (16)] hints at a possible mechanism of polymerization.