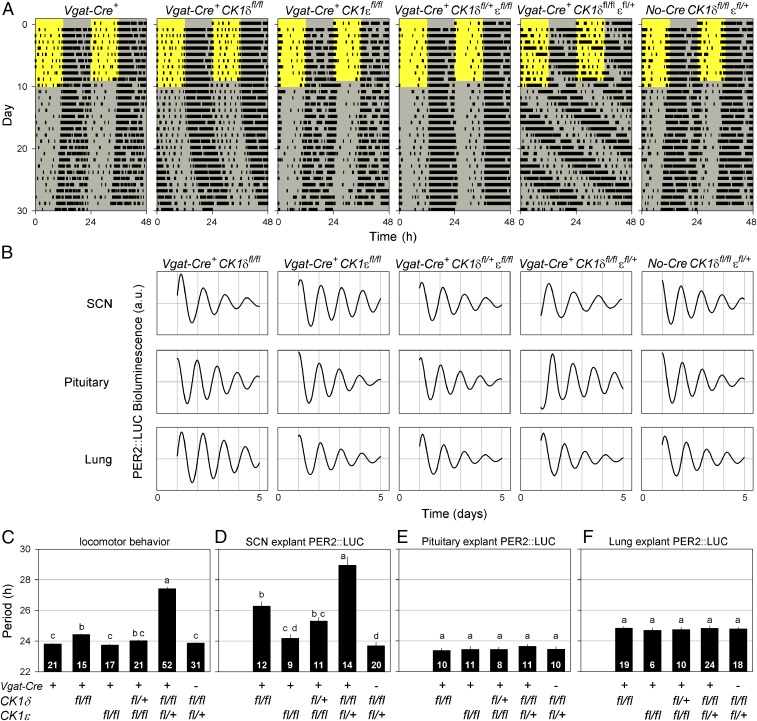
Fig. 1.
Disruption of CK1δ/ε alleles in GABAergic cells alters the period of behavioral and SCN rhythms but not peripheral clocks. (A) Representative, double-plotted wheel-running actograms of different Vgat-Cre+ CK1δ/ε genotypes illustrate the role of CK1δ/ε in setting the speed of the circadian clock. Mice were housed in a 12-h light/12-h dark lighting cycle for 10 d, followed by 20 d in constant darkness. The light and dark phases are indicated by the yellow and gray background, respectively. (B) Representative PER2::LUC bioluminescence traces of SCN, pituitary, and lung explants for different genotypes. High-amplitude oscillations in all genotypes show that disruption of CK1δ/ε alleles does not impair molecular circadian oscillations. (C–F) Period comparisons between different Vgat-Cre CK1δ/ε genotypes of locomotor behavior (C) and PER2::LUC bioluminescence rhythms of SCN (D), pituitary (E), and lung explants (F). Deletion of CK1δ/ε alleles in GABAergic cells results in alterations in locomotor and SCN period but does not affect the endogenous period of peripheral clocks. Bar graphs are mean ± SEM. The number of animals per genotype is indicated at the base of each bar. Genotypes identified with different letters are significantly different.