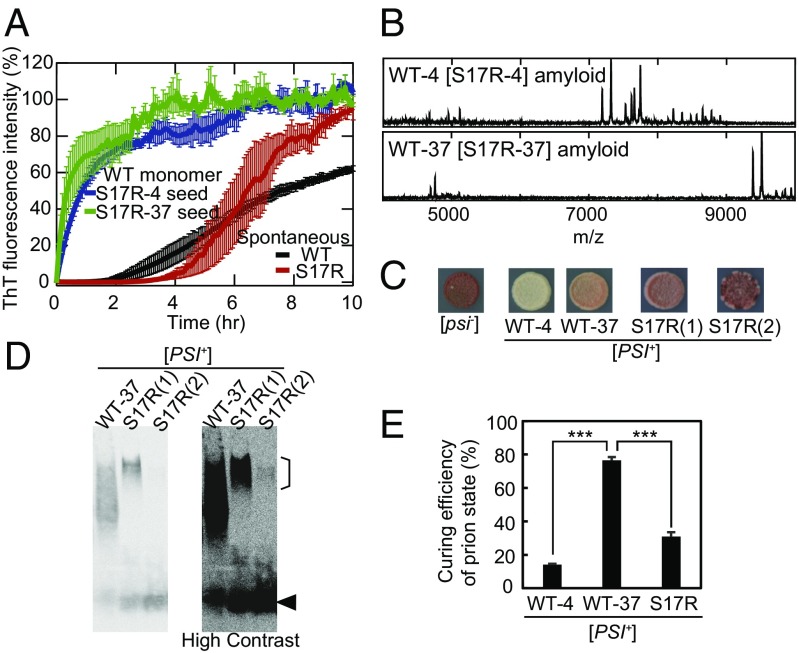
Fig. 2.
Wild-type Sup35NM can adopt the PrD-C amyloid core. (A) Kinetics of spontaneous WT and S17R amyloid formation and seeding reaction with WT monomer and S17R-4 and S17R-37 seeds monitored by ThT fluorescence at 25 °C. Error bars denote SEM. (B) High-m/z-range MALDI-TOF-MS spectra (m/z 4,000–10,000) of core peptides derived from cross-seeded WT[S17R] amyloid formed at 4 °C (WT-4[S17R-4]) and 37 °C (WT-37[S17R-37]). (C) Weak/sectoring phenotypes of two examples of [PSI+(S17R)] strains obtained by overexpression of the Sup35NM S17R mutant in [psi−] yeast. Phenotypes of [PSI+(WT-4)] and [PSI+(WT-37)] strains and [psi−] yeast are also shown as controls. (D) SDD-AGE analysis with yeast lysates of [PSI+(WT-37)] and [PSI+(S17R)] strains probed by an anti-Sup35NM antibody. A high-contrast image is shown on the right. An arrowhead and a line in the high molecular region indicate the positions of monomeric Sup35NM and S17R-type prions, respectively. Note that [PSI+(S17R (2))] strains also contain a large size of prion aggregates. (E) Efficiency of [PSI+]-state curing of [PSI+(WT-4)], [PSI+(WT-37)], and [PSI+(S17R)] strains by Hsp104 overexpression. [PSI+(S17R)] strains were generated by in vitro infection of Sup35NM S17R-4 or S17R-37 amyloid fibers in [psi−] yeast. The curing efficiency of [PSI+(S17R)] is an average of the data from the two [PSI+(S17R)] strains. Error bars denote SEM. ***P < 0.001 by a Student’s t test; n ≥ 3.