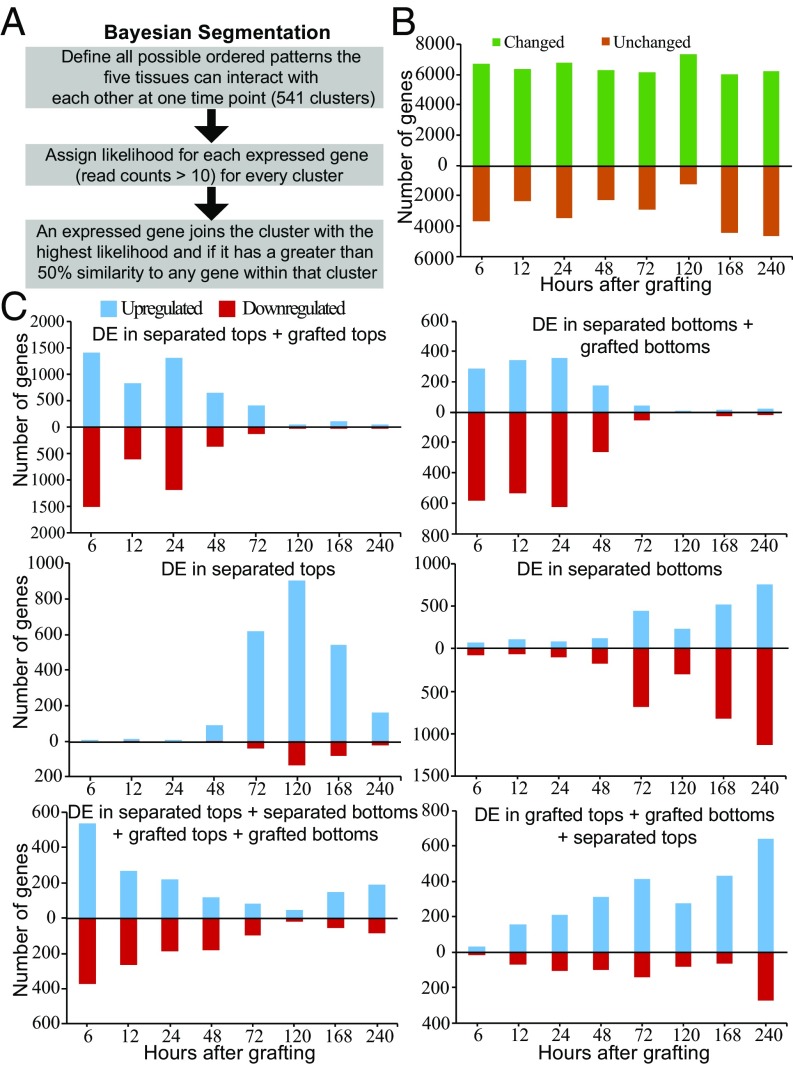
Fig. 6.
Clustering the transcriptome at each time point, based on likelihoods of all possible patterns of differential expression (DE) in grafted, separated, and intact tissues. (A) Overview depicting the Bayesian segmentation. (B) Analysis of differential behavior produced 113 categories containing at least 10 genes, the expression of which was in a specific differential pattern for at least one time point (Dataset S2). One group is composed of genes the transcript levels of which are not substantially changed in the five tissues (unchanged), whereas the other group is composed of the sum of the other 112 groups (genes the transcript levels of which changed after treatment in at least one tissue) over the time points tested. (C) Major categories in the segmentation revealed RNAs the levels of which changed in all of the treatments listed relative to intact samples. Note that a gene can be represented in only one category for a given time point, that is, the category in which the transcript level changes best fit the category.