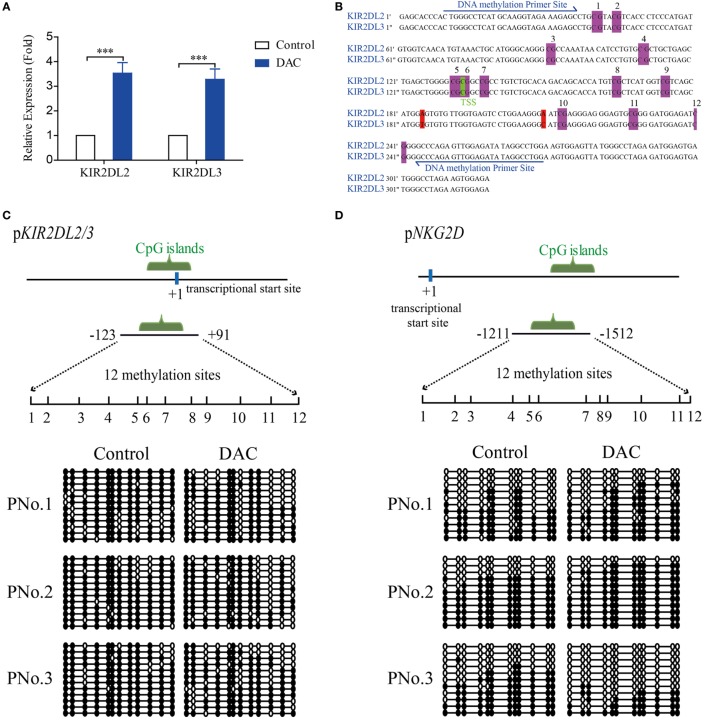
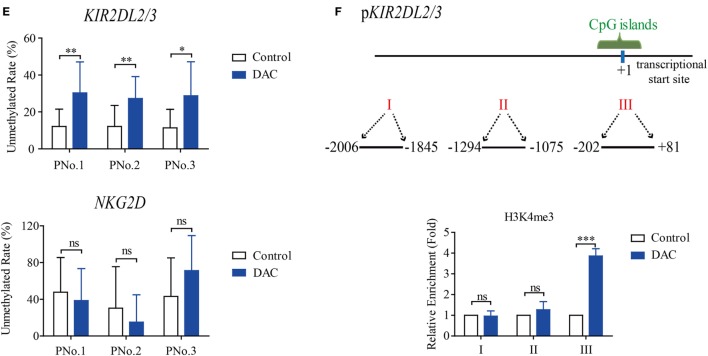
Figure 4.
Decitabine increased demethylation of KIR2DL2/3 promoter, which was beneficial to the transcription of KIR2DL2/3 in gamma delta (γδ) T cells. (A) qRT-PCR analysis of KIR2DL2 and KIR2DL3 expression on γδ T cells exposed to 0.5 µM DAC for 48 h. Data are expressed as the expression of KIR2DL2 and KIR2DL3 mRNA on DAC-treated γδ T cells relative to that on γδ T cells (***p < 0.001; n = 3). (B) The genes around the transcriptional star sites (TSS) (marked green) of KIR2DL2 and KIR2DL3 are nearly the same, except for two bases (marked red). There are 12 CpG sites (marked purple) around the TSS of KIR2DL2 and KIR2DL3 genes. (C,D) DNA methylation of the promoters of both KIR2DL2/3 and NKG2D in γδ T cells. The upper images show the schematic diagram of CpG islands in KIR2DL2/3 and NKG2D promoters. Vertical lines: methylation sites. The lower images show DNA methylation of KIR2DL2/3 and NKG2D promoters in γδ T cells treated with and without (control) DAC for 48 h from three patients. Open circles: unmethylated CpGs; solid circles: methylated CpGs. (E) Statistical analysis of the demethylation states of KIR2DL2/3 and NKG2D promoters in γδ T cells from three patients (*p < 0.05; **p < 0.01; ns, not significant). (F) Three regions (I, II, and III) including TSS (+1) of KIR2DL2/3 were analyzed by chromatin immunoprecipitation qPCR for H3K4me3 occupancy in the KIR2DL2/3 promoter of γδ T cells treated with and without (control) DAC for 48 h. Enrichment of KIR2DL2/3 promoter-specific DNA sequences was measured using quantitative PCR. Data are shown from three independent experiments and each PCR was performed in triplicate (***p < 0.001; ns, not significant).