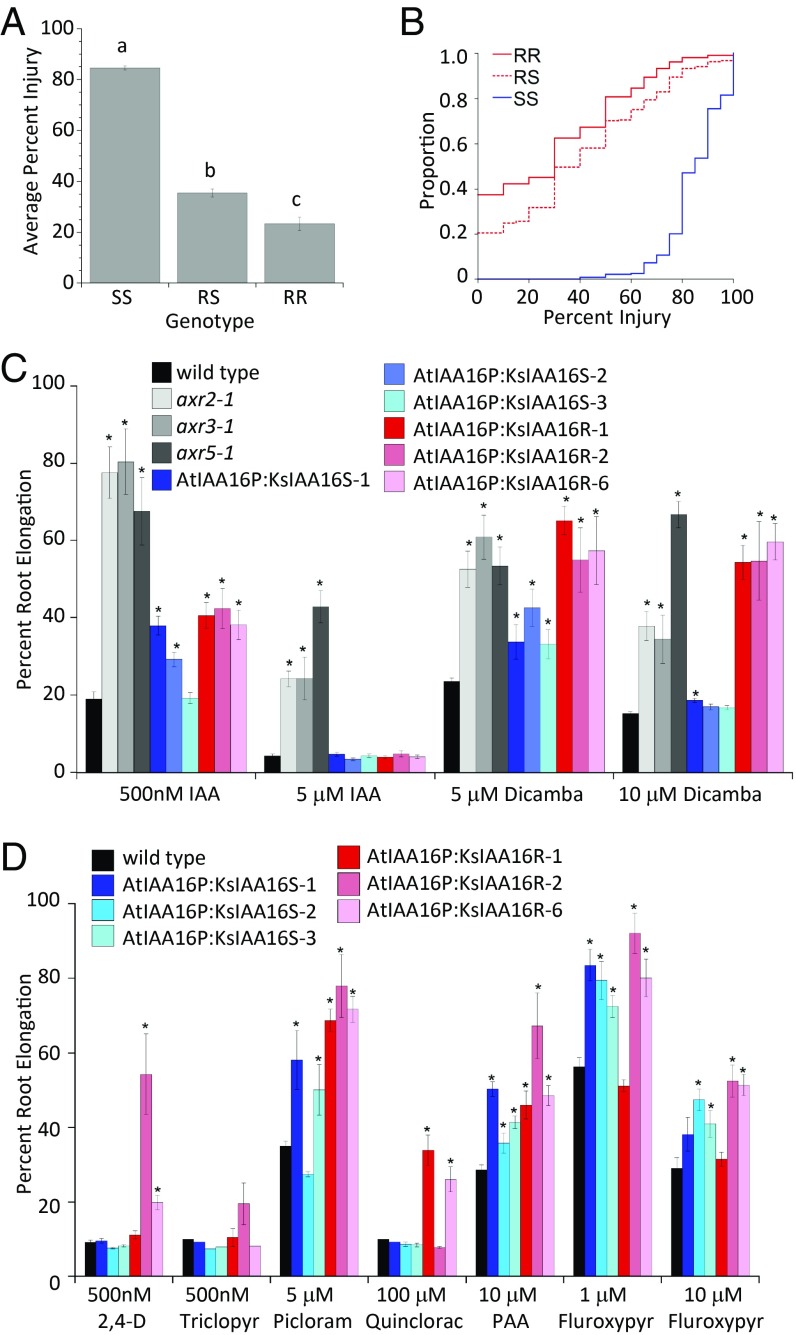
Fig. 4.
The KsIAA16 R allele confers dicamba resistance in kochia and Arabidopsis. A shows average injury by genotype of the segregating F2 kochia population and different letters a, b, and c indicate values that are significantly different from one another; 710 F2 progeny from three independently verified F1 parental plants were sampled, successfully genotyped, and sprayed with dicamba at a 1,120-g ha−1 rate. Based on the TaqMan genotyping assay, n = 234 for SS individuals, n = 371 for RS, and n = 105 for RR. B shows a Cramér–von Mises plot of injury by genotype for this same kochia experiment, which indicates that the R allele is semidominant. C shows the root elongation inhibition by 5 and 10 μM dicamba and 500 nM and 5 μM IAA in three independent transgenic Arabidopsis lines carrying either the KsIAA16S allele (blue shades) or the KsIAA16R (red shades) as well as the Arabidopsis mutants axr2-1, axr3-1, and axr5-1 (gray shades) for comparison. Each column represents the mean of 12 individuals, error bars represent SEs, and asterisks denote significant difference from the wild type. *P < 0.05. D shows the percentage of root elongation of these same transgenic Arabidopsis lines on other auxins, including 500 nM 2,4-D, 500 nM triclopyr, 5 μM picloram, 100 μM quinclorac, 10 μM PAA, and 1 and 10 μM fluroxypyr. Each column represents the mean of 12 individuals, error bars represent SEs, and asterisks denote significant difference from the wild type. *P < 0.05.