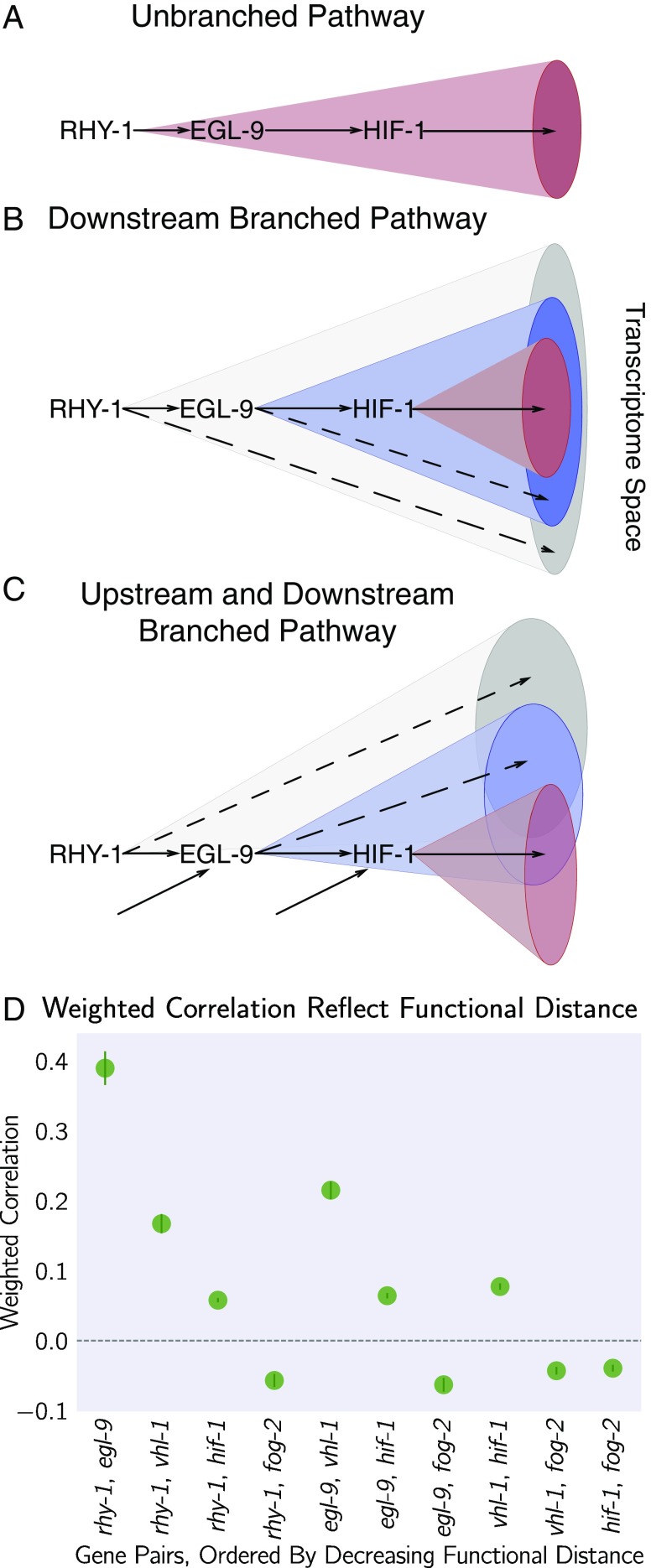
Fig. 6.
Transcriptomes can be used to order genes in a pathway under certain assumptions. Arrows in the diagrams above are intended to show the direction of flow and do not indicate valence. (A) A linear pathway in which rhy-1 is the only gene controlling egl-9, which in turn controls hif-1, does not contain information to infer the order between genes. (B) If rhy-1 and egl-9 have transcriptomic effects that are separable from hif-1, then the rhy-1 transcriptome should contain contributions from egl-9, hif-1 and egl-9– and hif-1–independent pathways. This pathway contains enough information to infer order. (C) If a pathway is branched both upstream and downstream, transcriptomes will show even faster decorrelation. Nodes that are separated by many edges may begin to behave almost independently of each other with marginal transcriptomic overlap or correlation. (D) The hypoxia pathway can be ordered. We hypothesize the rapid decay in correlation is due to a mixture of upstream and downstream branching that happens along this pathway. Bars show the SE of the weighted coefficient from the Monte Carlo Markov Chain computations.