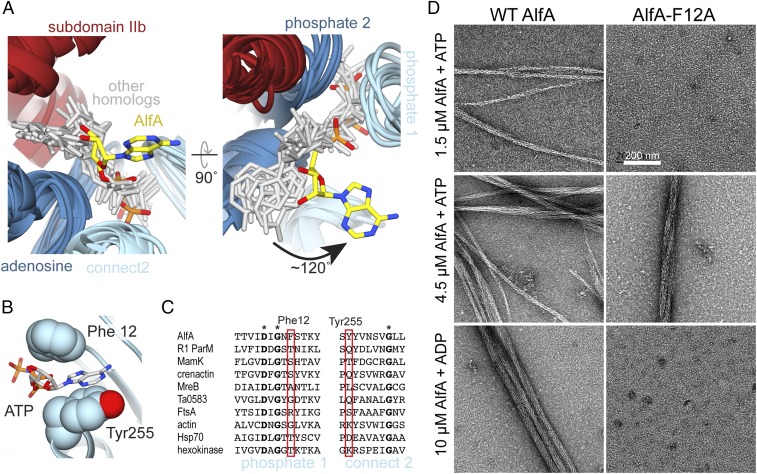
Fig. 3.
AlfA binds ATP through novel interactions. (A) Structural alignment of ATP binding sites of AlfA and other homologs of the actin/Hsp70/sugar kinase family bound to nucleotide, with the proteins rendered as ribbons (colored by subdomain as in Fig. 1), and nucleotides rendered as sticks (gray and yellow). The aligned structures are from cryo-EM filament reconstructions (PDB ID codes: actin, 5JLF; MamK, 5JLV; R1 ParM, 5AEY; crenactin, 5MW1), and crystal structures (PDB ID codes: MreB, 4CZJ; Ta0583, 2FSN; FtsA, 1E4G; Hsp70, 3KVG; hexokinase, 2E2Q). Structural alignment was performed using just the regions around the conserved actin sequence motifs. (B) In the AlfA filament structure the adenosine base is sandwiched between Phe12 and Tyr255 in subdomain Ia. (C) Sequence alignment of the phosphate 1 and connect 2 actin motifs, with positions of Phe12 and Tyr255 highlighted in red. Invariant positions are marked with an asterisk. (D) Negative-stain electron micrographs of AlfA wild-type and F12A mutant in the presence of ATP and ADP. The F12A mutation is capable of assembling filaments but cannot maintain stable filaments in ADP.