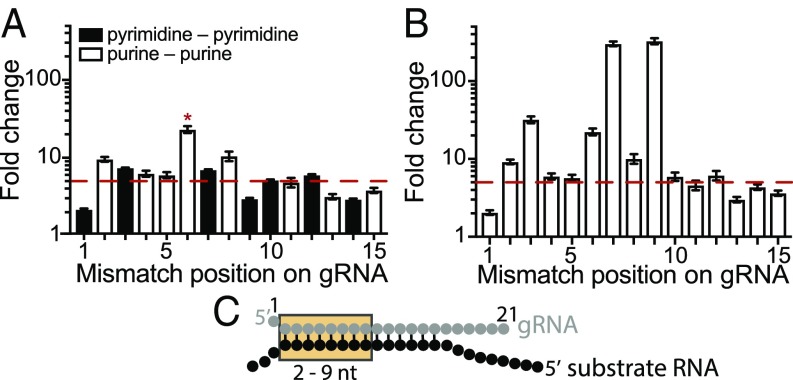
Fig. 4.
Determination of the gRNA seed region of MpAgo RNPs. (A) Substrate ssRNA filter-binding assays with ssRNA substrates that contain a single-nucleotide mismatch along the gRNA from the first to 15th position. The sequence of the gRNA was kept constant, and the ratios of the average Kds of the substrates that create mismatch (Kdmm) to the fully complementary substrate (KdCompl; fold change = Kdmm/KdCompl) were plotted against the position of the mismatch. The red dash line indicates the value equal to ∼5 times the Kd for the fully complementary ssRNA substrate. Asterisk (*), A–G mismatch at the sixth position (guide RNA–substrate RNA). The Kd for a G–A mismatch at the sixth position is 779 ± 71 pM, using a different gRNA (SI Appendix, Table S1). (B) The ratios of the average Kds of the substrates that create purine–purine mismatch (Kdmm) to the fully complementary substrate (KdCompl; fold change = Kdmm/KdCompl) were plotted against the position of the mismatch. The red dashed line indicates a value equal to 5. The purine–purine mismatches at the second through 12th positions decrease the affinity of MpAgo RNP more than fivefold. The third, sixth, seventh, and ninth positions reduce the affinity of MpAgo RNP the most: 32-, 22-, 301-, and 327-fold respectively. (C) The gRNA-substrate RNA duplex with the seed region (orange box).