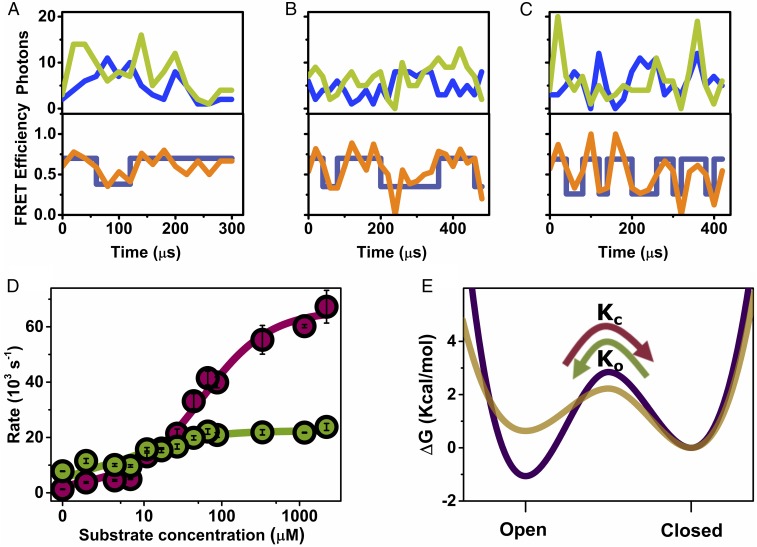
Fig. 2.
Domain closure dynamics under turnover conditions. (A–C) smFRET trajectories measured at increasing substrate concentration (ATP + ADP; Table S1): 4.2 μM (A), 16.8 μM (B), and 1.16 mM (C), with AMP at a fixed concentration of 1 mM. Top in each part shows donor (blue) and acceptor (light green) signals binned in 20 µs bins, and the Bottom shows the calculated FRET efficiency (orange), and the state of the system at each time bin, as calculated by the Viterbi algorithm based on the H2MM analysis (dark blue). (D) Closing and opening rates (cherry and green circles, respectively) as a function of substrate concentration, obtained from H2MM analysis of a series of smFRET experiments. Error bars represent standard errors of the mean. Continuous lines are fits to the model described in the text, from which closing and opening rate constants for the unbound and bound states of the enzyme were extracted. (E) Free energy profiles for the unbound (purple) and bound (gold) enzyme, calculated from the fitted rates (with an assumed preexponential factor of 1 μs in the Kramers rate expression) (38), indicate that the open conformation is more stable than the closed conformation in the unbound state, with the situation inverted in the bound state. The transition state free energy is reduced in the bound state, leading to faster domain closure dynamics. The curvatures and shapes of the free energy profiles were selected for visualization purposes only.