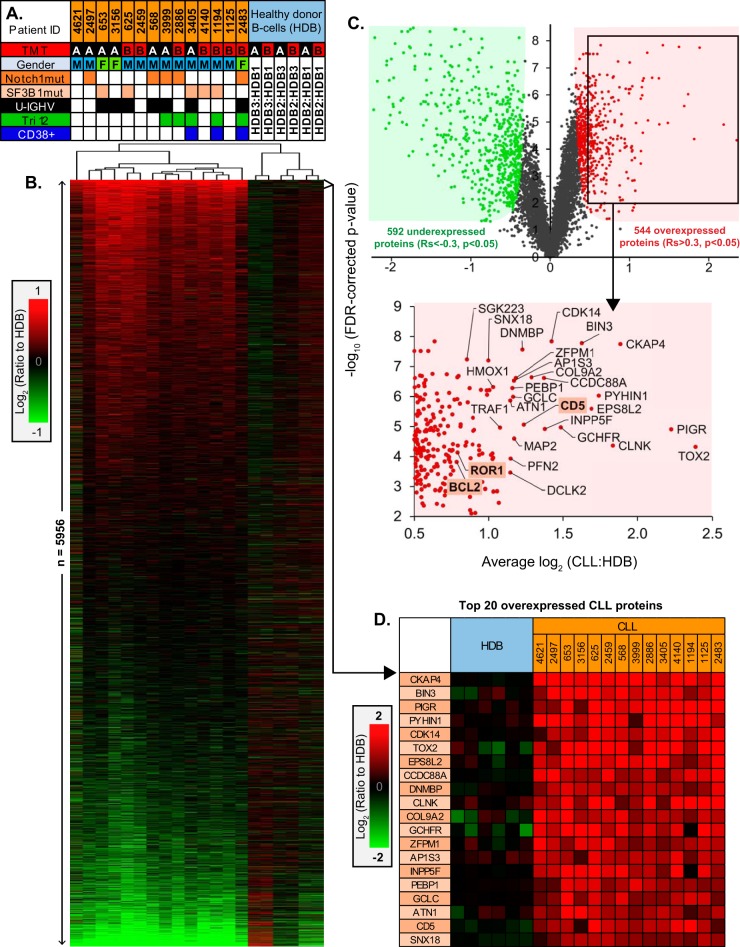
Fig. 2.
Quantitative proteomics profiles and characteristics of CLL samples. A, Summary of the major characteristics of the 14 CLL samples, including; the assigned TMT 10-plex experiment (A or B), patient gender, presence of mutations to either NOTCH1 or SF3B1 genes, IGHV mutation status (U-IGHV, solid fill; unmutated IGHV, open), trisomy 12 status (Tri12), CD38+ (>99%) cases. B, Hierarchical clustering of 5956 protein log2 (ratios) for the 14 CLL samples relative to HDB, sorted by regulation score. C, Volcano plot demonstrating proteins observed with significant overexpression (Rs>0.3, p < 0.05) or underexpression (Rs<-0.3, p < 0.05) in CLL versus HDB. A selection of the most overexpressed proteins are annotated, identifying the accepted CLL-specific hallmarks BCL2, ROR1 and CD5. D, The top 20 most consistently overexpressed proteins in CLL identified by the proteomics.