Fig. 13.
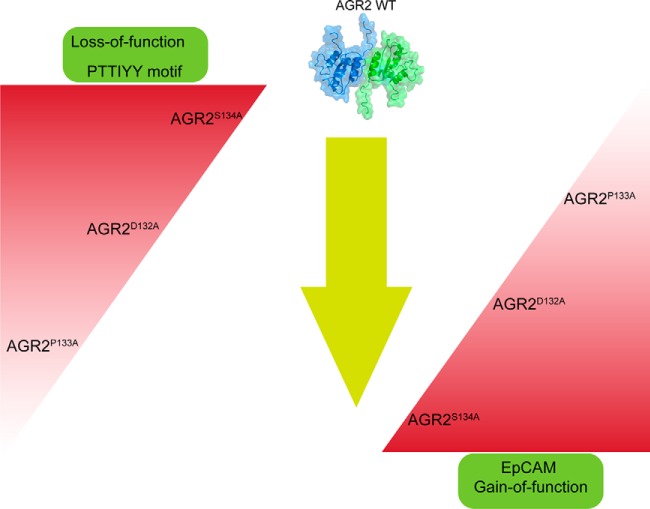
Summary of the biochemical properties of the peptide-docking site mutations in AGR2 produced based on hydrogen-deuterium exchange mapping. Based on the hydrogen-deuterium exchange mapping data (Fig. 4 and 5), we focused on creating three alanine substitutions mutations in the VDPSL loop motif, from amino acids 131–135, residing between two β-sheets (Fig. 6). The mutant proteins exhibit inverse trends in their specific activity. Consensus peptide binding reactions demonstrate that wt-AGR2 = S134A>D132A>P133A with two mutants showing a loss-of-function. Although in EpCAM binding, S134A>D132A>P133A = wt-AGR2 with two mutants showing a gain-of-function. These data suggest that although mutating some amino acids in the VDPSL motif can impact on specific peptide binding, the global conformation changed induced by loop mutation (for example in the gain-of-function S134A mutation (Fig. 11) might result in binding to a distinct site on the EpCAM molecule. Thus, one interpretation of such data is that one purpose of the VDPSL motif is to not only drive specific peptide binding by AGR2 but to constrain the conformational dynamics (or monomer-dimer equilibrium) of AGR2 so as to minimize its “binding to other sites” on its client proteins.