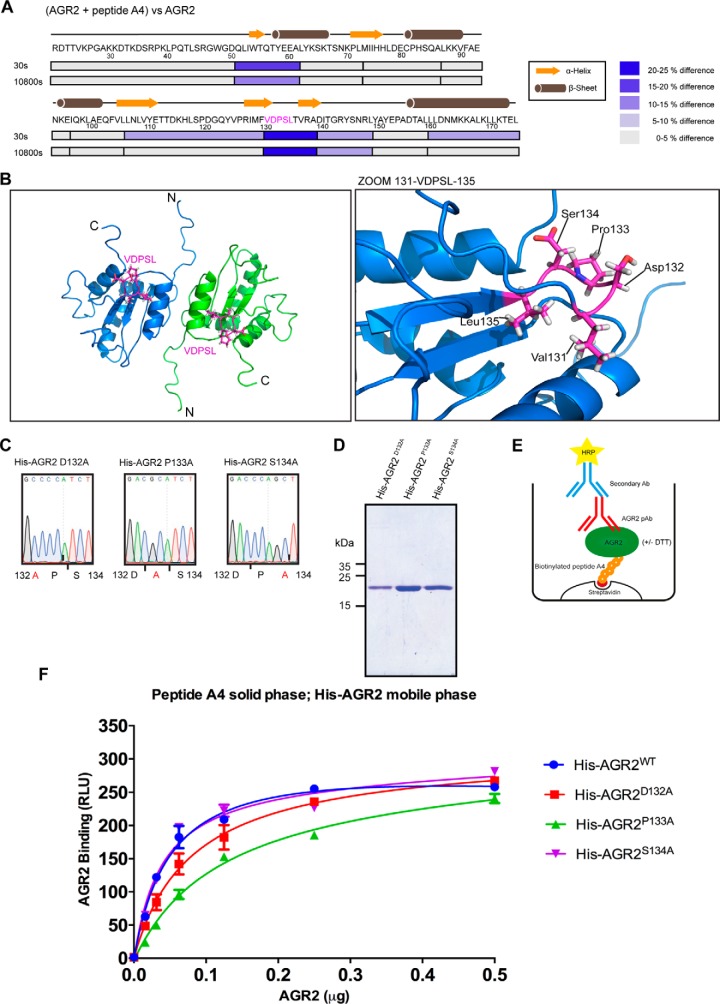
Fig. 6.
Mutations in the dominant deuteration responsive motif impact on AGR2 peptide binding activity in vitro. A, A secondary structure summary of changes in deuteration in the presence of the consensus peptide. The diagram shows the full amino acid sequence of the mature AGR2 protein (amino acids 21–175) with the alpha helices and β-sheets highlighted. The block colors highlight the changes in fold deuteration after 30 s or 10800 s, with dominant changes at the region 131–139 (in between two β-sheets) and the dimer interface (a 50–60). B, A representation of the minimal deuteration responsive motif from amino acids 131–135 motif (highlighted in pink) in AGR2 (PDB code: 2LNS) that is most significantly suppressed by peptide A4 binding. The main amino acids of focus were D132, P133, and S134 flanked by the hydrophobic amino acids V131 and L135. C, Data showing alanine substitution mutations generated at codons D132, P133, and S134 in bacterial expression plasmids with the corresponding DNA sequencing chromatogram traces of three of AGR2 peptide docking site mutations at position Asp132 to Ala, Pro133 to Ala, and Ser134 to Ala. D, An SDS-Coomassie blue gel showing the relative purity of the indicated mutant proteins expressed in E. coli after nickel affinity purification. E–F, An ELISA assay was developed to measure the binding of AGR2 to synthetic biotinylated peptide A4 captured on the streptavidin coated solid phase. Reactions were added to the solid phase to measure binding to biotinylated peptide A4 on the solid phase. The data plot the binding of AGR2 in relative light units (RLU) as a function of increasing wt or mutant AGR2 protein isoforms, as indicate (in μg) that was quantified using AGR2 specific antibody.