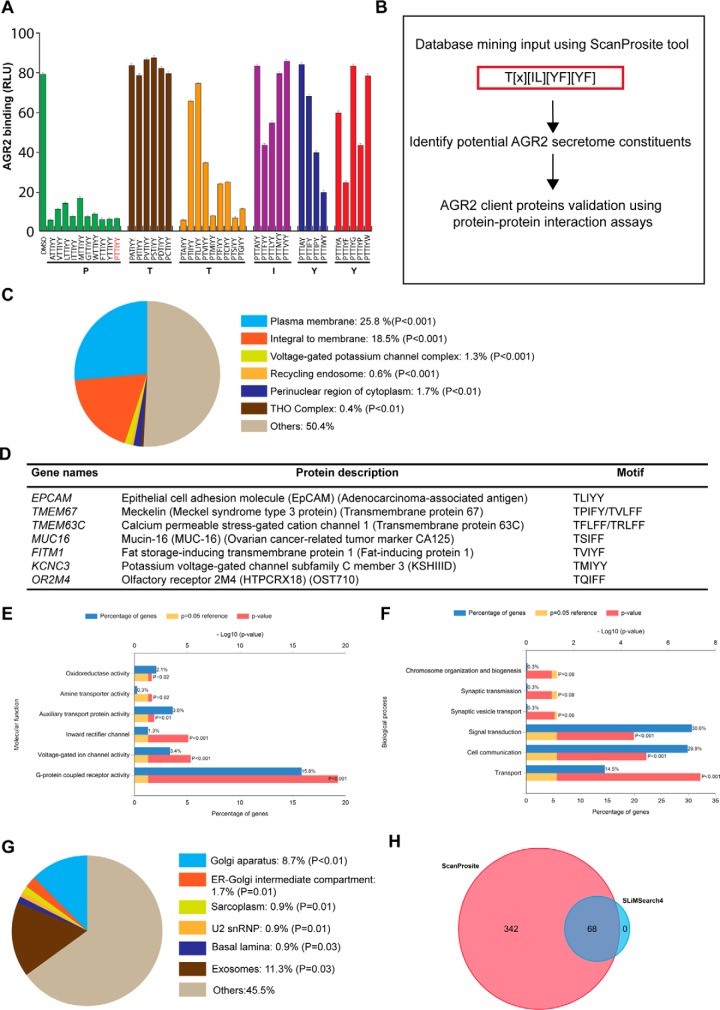
Fig. 7.
Mining of the human proteome for proteins containing the AGR2 linear peptide consensus motif. A, The PTTIYY hexapeptide was previously defined as a minimal peptide sequence that binds to AGR2 (24). A mutational scan library was synthesized containing a subset of amino acid substitutions at positions 1–6, from left PTTIYY. The substitutions included small hydrophobic (L, V, I, M, A, G), bulky hydrophobic (W, F, P), charged (D, C) and hydrophilic (S, T) of the linear peptide motif. The peptides sequences created are shown on the X-axis. The biotinylated peptide A4 was bound to the streptavidin-coated solid phase and fixed amounts (1 μg) of AGR2 protein with 100 ng of the indicated synthetic peptide was added. AGR2 binding was detected using a secondary antibody and binding is measured in as RLU. The data revealed that amino acids at positions 2, 4, 5, and 6 are relatively fixed whereas changes at positions 1 and 3 can be relatively well tolerated. B, Schematic illustrating strategy to find novel AGR2 client proteins using linear peptide motif database mining. The AGR2 linear peptide consensus motif was used as input using a ScanProsite tool (http://prosite.expasy.org/scanprosite/) and the human proteome database was screened to identify proteins containing the motif. C, The scan resulted in 409 protein hits when splice variants were excluded (supplemental Tables S1 and S2). The hits were scored as subcellular localization using FunRich (v2.1.2) (62) where the majority of the proteins found were membrane proteins. A large proportion of the hits were membrane-related proteins, which foreshadows AGR2 function in receptor maturation. Enriched terms were ranked by p value (Hypergeometric test). D, Representative of possible AGR2 binding proteins is shown containing the consensus peptide-binding motif. E, Bar graph of molecular function overrepresented in AGR2 linear peptide motif hits. F, Bar graph of biological processes overrepresented in AGR2 linear peptide motif hits (Supplemental Tables 1 and 2). The percentage of genes linking to the individual enriched terms were ranked by p value and are shown together with the p value from the Hypergeometric test (depicted in red) and the reference p = 0.05 value (depicted in yellow). G, The AGR2 linear peptide consensus motif was used as input using the SLiMSEARCH4 linear motif discovery tool (http://slim.ucd.ie/slimsearch/index.php) and the human proteome database was screened to identify proteins containing the motif. Enriched terms were ranked by p value (Hypergeometric test). H, Venn diagram highlighting the overlap and number of linear motifs identified between ScanProsite and SLiMSEARCH4 tools.