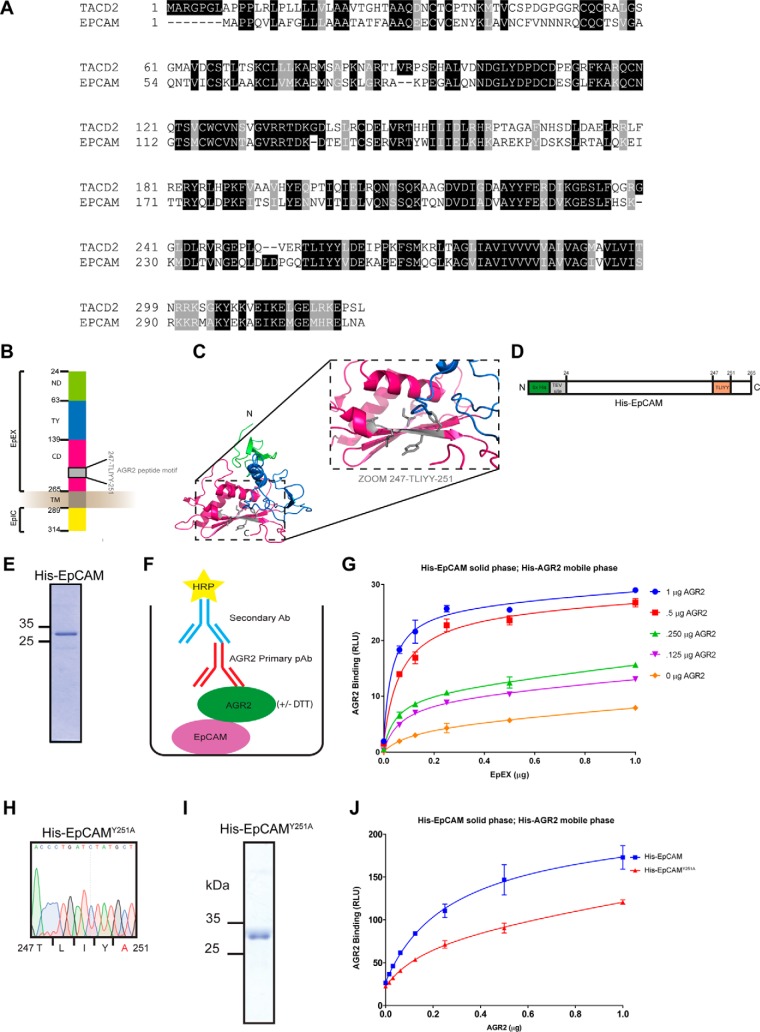
Fig. 8.
EpCAM as a candidate AGR2 client protein. A, Homology between EpCAM and its paralogue TACD2 as aligned using Clustal Omega. Both proteins were identified using ScanProsite (supplemental Table S1) and harbor the TLIYY motif implicated as an AGR2 linear peptide docking site. B, Secondary structure of EpCAM which consists of a N-domain (ND, green), Thyroglobulin type-1 domain (TY, blue) and C-domain (CD, dark pink) which altogether make up for extracellular domain (EpEX), transmembrane domain (TM, gray), intracellular domain (EpIC, yellow), and the amino acids from 247–251 containing the sequence TLIYY. C, Three-dimensional cartoon representation of extracellular part of human EpCAM (PDB code: 4MZ) highlighting the AGR2 linear peptide motif at amino acid position Thr247 to Tyr251 9 (gray). Color coding is the same as in (B). D, Schematic representation of his-tagged EpCAM protein sequence highlighting the extracellular domain (EpEX), the TEV cleavage site, and the TLIYY motif. E, Coomassie Blue staining of purified His-EpCAM showing the major band at 32 kDa under reducing SDS-PAGE condition. F–G, Solid-phase binding assay to measure AGR2 binding to EpCAM protein. Increasing amounts of EpCAM were immobilized on the surface of a microtiter plate (0–1 μg). AGR2 (0–1 μg) was titrated in the mobile phase and AGR2 binding to immobilized EpCAM was quantified using AGR2 specific antibody. The binding of AGR2 is plotted as the extent of protein-protein complex formation as RLU as a function of increasing protein in the mobile phase. H, DNA sequencing chromatogram traces of EpCAMY251A. I, Coomassie Blue staining of purified His- EpCAMY251A mutant protein showing the major band at 32 kDa under denaturing SDS-PAGE. J, His-EpCAM or His-EpCAMY251A (1 μg) was immobilized onto the well surface of a microtiter plate as in (F) His-AGR2 WT (0–1 μg) was titrated in the mobile phase. AGR2 binding to immobilized EpCAM was quantified using AGR2 specific antibody. The binding is plotted as the extent of protein-protein complex formation as RLU as a function of increasing protein in the mobile phase.