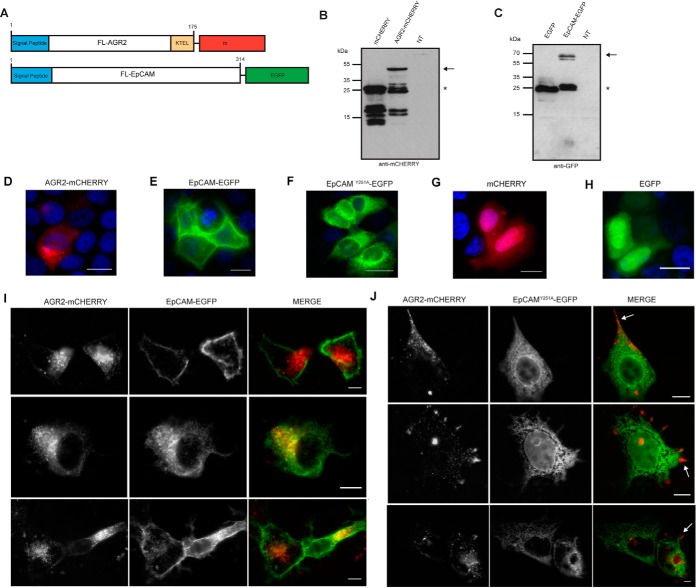
Fig. 9.
Effects of Y251A mutation on EpCAM localization in cells. A–C, Fluorescently labeled versions of AGR2 (mCHERRY) and EpCAM (EGFP) with the signal peptides were generated and expression validated in cells using immunoblotting (B and C). B, The immunoblotting of mCHERRY and mCHERRY-AGR2 transfected cells highlights the expression of mCHERRY alone (lane 1) and mCHERRY-AGR2 (lane 2). Blots were incubated with an anti-mCHERRY antibody. The arrow marks the location of full-length mCHERRY-AGR2 and the asterisk marks the location of mCHERRY. We noticed the reproducible small molecular mass “cleavage” or synthesis products when mCHERRY was transfected into cells. C, The immunoblotting of EGFP and EGFP-EpCAM transfected cells highlights the expression of EGFP alone (lane 1) and EGFP-EpCAM (lane 2). Blots were incubated with a GFP antibody. The arrow marks the location of full-length EGFP-EpCAM and the asterisk mark the location of EGFP. The EGFP was not subjected to the production of smaller molecular mass adducts as was the mCHERRY protein. Fluorescent microscopy was used to measure the relative localization of the following proteins; D, mCHERRY-AGR2 E, EGFP-EpCAM F, EGFP-EpCAMY251A G, mCHERRY and H, EGFP. I and J, The impact of cotransfection of mCHERRY-AGR2 and EGFP-EpCAM or EGFP-EpCAMY251A on their respective localizations. Representative images of the wild-type and mutant EpCAM, as well as AGR2, are highlighted in both panels. The arrow in J highlights AGR2 mislocalization to the plasma membrane periphery in EpCAM mutant cotransfections that mirrors the mutant EpCAM mislocalization to the nuclear membrane in the same cells.