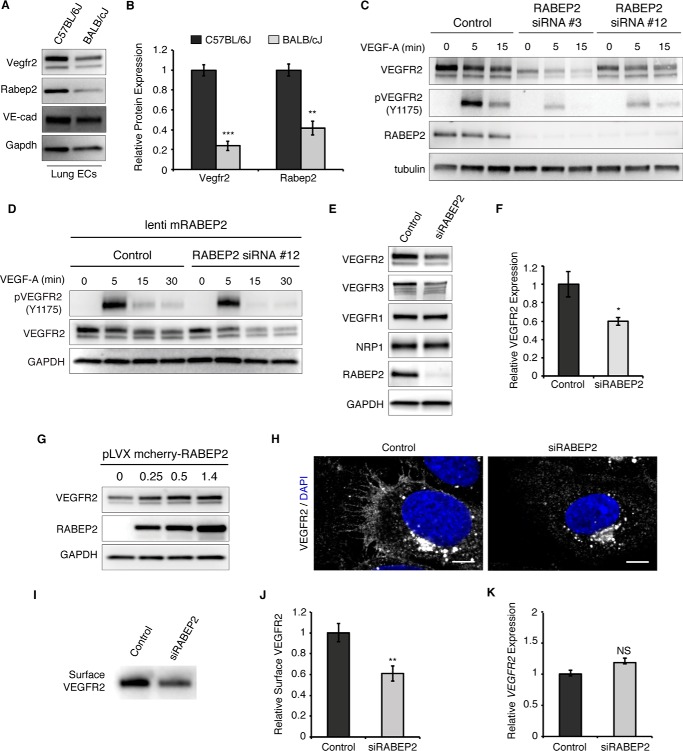
Figure 1.
RABEP2 regulates VEGFR2 expression. A, protein lysate isolated from either C57Bl/6J or BALB/cJ lung endothelial cells was probed for Vegfr2, Rabep2, and VE-cadherin (VE-cad) to validate endothelial cell identity. Gapdh served as a loading control. B, quantification of Western blot showing relative Vegfr2 and Rabep2 protein expression. Quantification based on 3 independent cell isolations (mean ± S.D., **, p < 0.01; ***, p < 0.01). C, validation of RABEP2 siRNA. HUVEC were exposed to two different siRNA with unique human RABEP2 targeting sequences (RABEP2 siRNA #3 and RABEP2 siRNA #12). Both siRNA constructs depleted RABEP2 expression and caused reduced VEGFR2 expression and activation. RABEP2 siRNA #12 was chosen for all future experiments, as it had the least impact on cell growth. D, specificity of RABEP2 siRNA #12 validated by showing lentiviral infection with mouse RABEP2 construct (mRABEP2) rescues VEGFR2 expression and activation. E, Western blot of protein lysates isolated from HUVEC transfected with scrambled siRNA (control) or siRNA targeting RABEP2 (siRABEP2). F, quantification of the relative total VEGFR2 protein expression based on 3 independent experiments (mean ± S.D., *, p < 0.05). G, Western blot of total VEGFR2 expression following increasing levels of RABEP2 overexpression via lentiviral infection in HUVEC. H, immunocytochemistry of control or siRABEP2 HUVEC stained for VEGFR2 (white) and DAPI (blue) to mark cell nuclei. I, Western blot probed for VEGFR2 of total surface-biotinylated protein isolated from control or siRABEP2 HUVEC. J, quantification of relative surface VEGFR2 levels based on 3 independent surface biotinylation assays (mean ± S.D., **p < 0.01). K, relative VEGFR2 transcript levels in control and siRABEP2 HUVEC (mean ± S.D., NS, not significant).