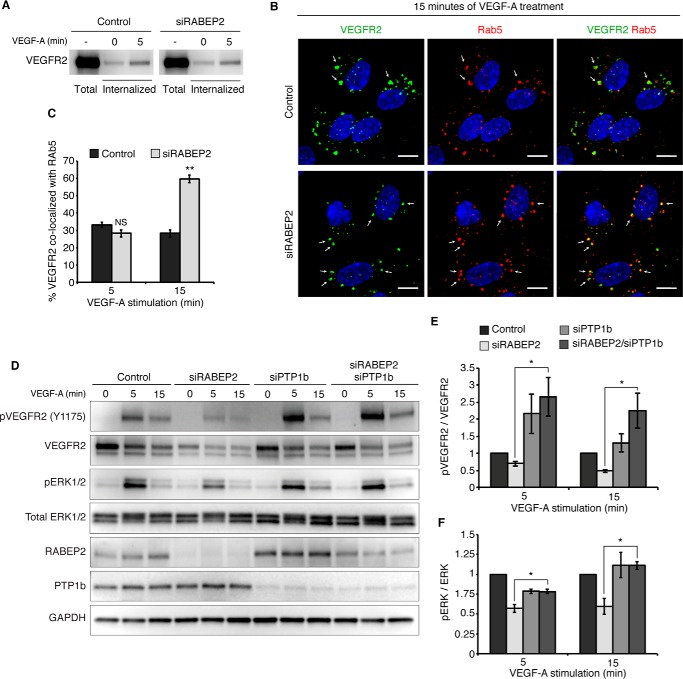
Figure 3.
RABEP2 regulates VEGFR2 endosomal trafficking. A, VEGFR2 internalization assay. Following incubation with biotin, cells were either harvested prior to stimulation to assess total surface VEGFR2 (total) or stimulated with 50 ng/ml of VEGF-A165 for 0 or 5 min (internalized), then stripped of the remaining surface biotin and lysed to assess levels of internalized biotin-labeled protein via Western blot probed for VEGFR2. B, HUVEC co-stained for internalized VEGFR2 (green) and Rab5 (red) after 15 min of stimulation with 50 ng/ml of VEGF-A165. White arrows point to Rab5+ endosomes containing internalized VEGFR2. DAPI marks cell nuclei in blue; scale bar, 10 μm. C, quantification of percent co-localization of internalized VEGFR2 and Rab5 after 5 and 15 min of VEGF-A165 stimulation. Quantification was performed using a co-localization plugin of Image J in at least 10 independent fields (mean ± S.D., NS, not significant; **, p < 0.01). D, HUVEC transfected with scrambled siRNA (control), scrambled siRNA and siRNA targeting RABEP2 (siRABEP2), scrambled siRNA and siRNA targeting PTP1b (siPTP), or siRNA targeting RABEP2 and PTP1b (siRABEP2/siPTP1b), serum starved for 12 h and then stimulated for 5 or 15 min with 50 ng/ml of VEGF-A165. Protein lysates were assessed by Western blots to assess VEGF signaling using antibodies against the tyrosine 1175 phosphorylation site of VEGFR2 (pVEGFR2 Y1175), total VEGFR2, RABEP2, and PTP1b to validate knockdown and GAPDH as a loading control. E, quantification of pVEGFR2 Tyr1175 normalized to total VEGFR2 of 3 independent experiments (mean ± S.D., *, p < 0.05). F, quantification of pERK1/2 normalized to total ERK1/2 of 3 independent experiments (mean ± S.D., *, p < 0.05).