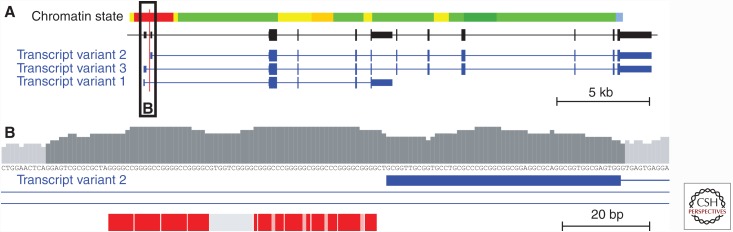
Figure 1.
Genomic organization of C9orf72 and the G4C2 hexanucleotide repeat at chromosome 9p21. (A) The C9orf72 gene and its major transcript variants numbered according to the National Center for Biotechnology Information (NCBI) RefSeq transcript collection. The red vertical line indicates the location of the noncoding G4C2 hexanucleotide repeat. The chromatin state illustrates transcriptional activity with the red bar indicating the active promoter based on combined analysis of histone marks in lymphoblastoid cell line GM12878 (Ernst et al. 2011). Black and blue boxes represent transcribed segments: wide boxes are coding, and narrow boxes are untranslated exonic regions. Horizontal connecting lines represent intronic regions. The black box labeled “B” is shown in detail in B. (B) Detailed view of the genomic context of hexanucleotide repeat sequence. The bar chart indicates relative 11 bp windowed GC content showing the contiguous GC-rich (>55%) low-complexity sequence in dark gray. Red boxes represent individual hexanucleotide repeat units in human reference genome sequence build GRCh37/hg19, with light red areas indicating nucleotide mismatches in the G4C2 consensus sequence. The gray box shows the common deletion in the 3′ flanking low-complexity sequence (LCS) containing the GTGGT motif, as frequently observed in expansion carriers. The blue box indicates the first noncoding exon of C9orf72 transcript variant 2, and blue lines indicate intronic regions.