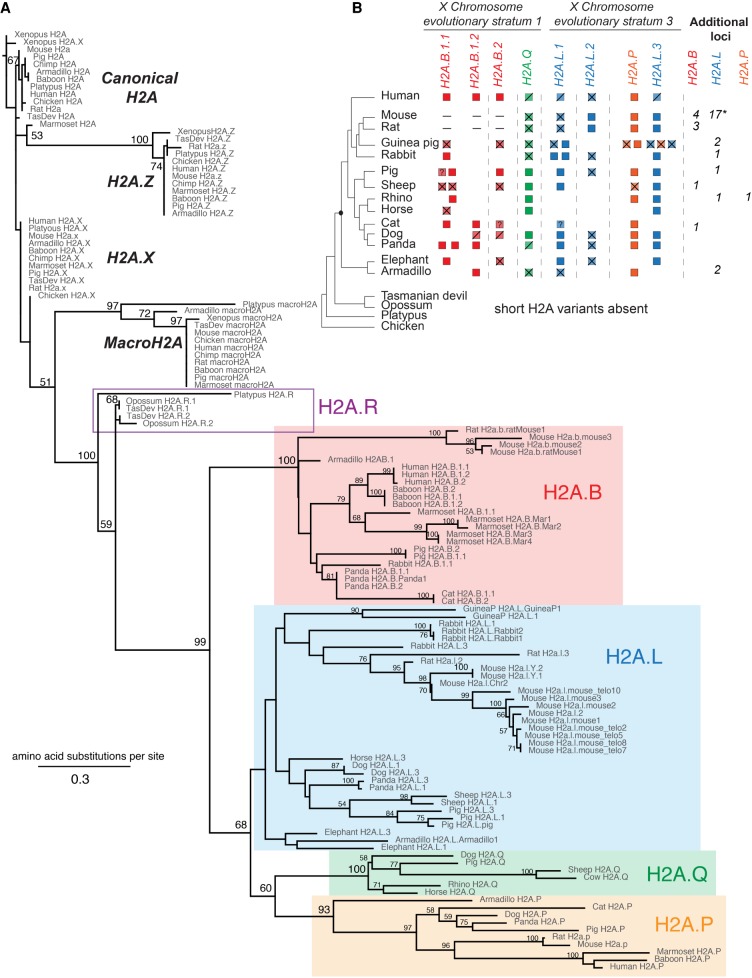
Figure 1.
Mammalian short H2A repertoires and phylogeny. (A) Maximum-likelihood protein phylogeny of the histone fold domain of canonical H2A and H2A variants from Xenopus, chicken, and representative mammalian species. Bootstrap values are shown at all nodes that have >50% bootstrap support. The tree is rooted using Xenopus H2A. Short H2A variant clades are highlighted: H2A.B (red box), H2A.P (orange box), H2A.Q (green box), and H2A.L (blue box). (B) Schematic representation of the shared syntenic locations encoding short H2A variants. Short H2As are indicated to the right of a species cladogram (Bininda-Emonds et al. 2007). The origin of the eutherian mammals is highlighted by a black dot. Intact open reading frames are shown with filled boxes. Disrupted open reading frames are shown with crossed boxes (confirmed pseudogenes) or diagonal lines (inferred pseudogenes) (see Results). A dash (–) indicates absence at the syntenic location, a “?” refers to incomplete sequence information, and an empty white space indicates a missing gene at that syntenic location—we have not attempted to distinguish true gene loss from absence due to assembly gaps; see Supplemental Figure S2 for details. X Chromosome evolutionary strata are indicated at the top (Lahn and Page 1999; Sandstedt and Tucker 2004; Ross et al. 2005). For the additional H2a.l loci in mouse, “*” denotes the presence of two Y-linked and one autosomal copies.