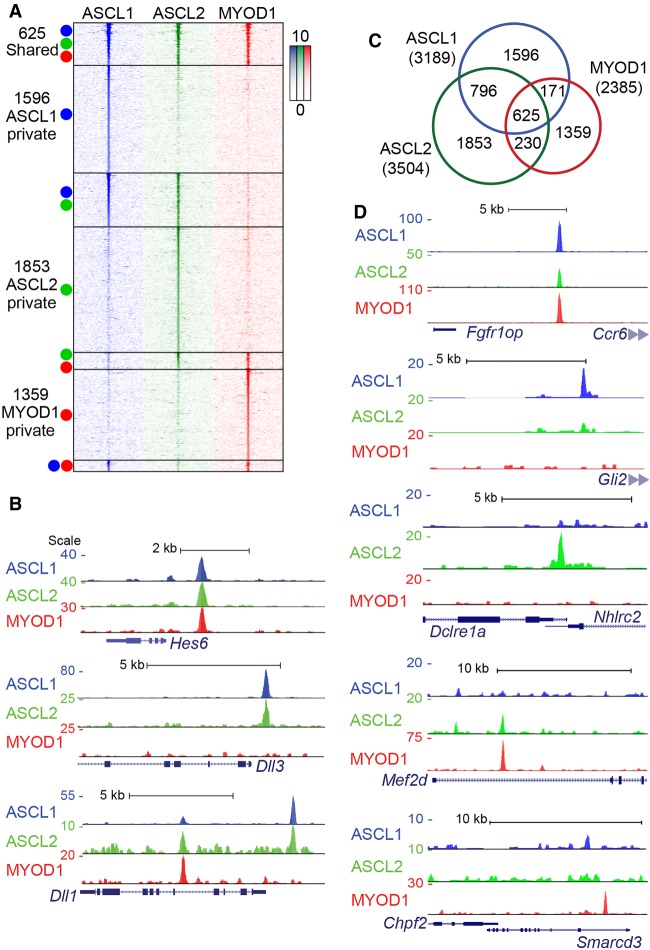
Figure 2.
Tissue-specific bHLH factors maintain distinct binding in the ESC genome revealing intrinsic binding specificity properties. (A) Heatmap of ChIP-seq data sets from ESCs at 24 h post-induction of bHLH expression. Each column shows the ChIP-seq signal for the factor indicated. Each row indicates a single interval ±3 kb centered on the peak apex, with the ChIP-seq signal indicated by color density. Colored circles indicate which ChIP-seq data set(s) identified a significant peak. (B,D) UCSC Genome Browser tracks comparing bHLH ChIP-seq enrichment near known targets including the Notch pathway genes Hes6, Dll3, and Dll1 (B), and novel targets (D) with either shared or private peaks. Each track shown represents ChIP-seq data from the indicated bHLH factor, normalized to 10 million reads and aligned to the mm10 genome. Track scale represents the number of reads at a given position. (C) Proportional area diagram of bHLH binding sites shown in the heatmap (A) with numbers representing distinct peaks identified in each subset. Peaks were considered overlapping if the peak apex was within 150 bp.