Figure 5.
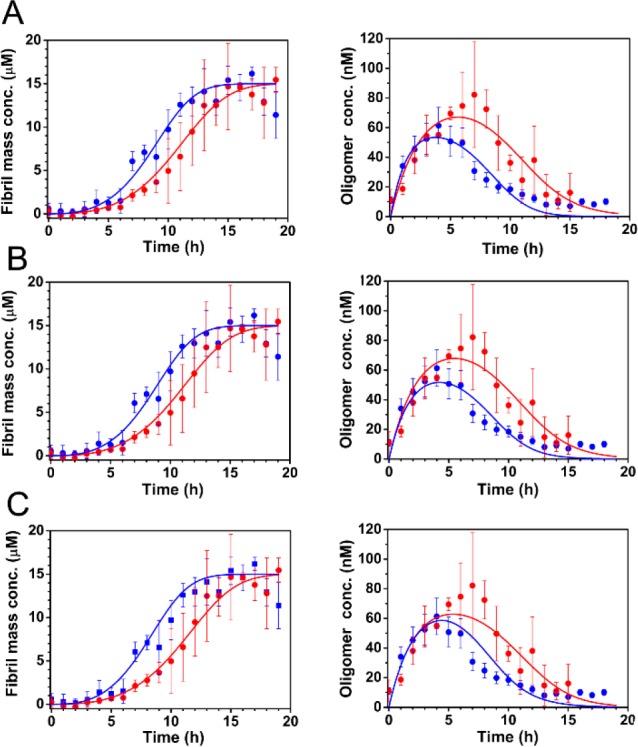
Fitting of combined smFRET/ThT data to models indicates a probable effect of mutations on the dissociation of oligomers, but not on their formation. (A–C) The bulk aggregation kinetics of 15 μM unlabeled Ure2-V9C (red) and Ure2-S68C (blue) monitored by ThT fluorescence (left panels) and the concentration of AF555/AF647 labeled Ure2-V9C (red) and Ure2-S68C (blue) oligomers throughout the aggregation reaction monitored by confocal smFRET (right panels) were globally fitted to a theoretical model (see Methods) including the formation, dissociation, and conversion of oligomers, and the elongation and fragmentation of fibrils. The incubation conditions were the same as in Figure 1. (A) Allowing both koligo and kd to differ for each mutant gives good fits, with a mean squared error of 1.54. (B) If koligo is constrained to be the same for both mutants, the model fits the data equally well, with a mean squared error of 1.58. (C) If neither koligo nor kd is allowed to differ, the fit is less good, especially around the time when the oligomer concentration is at a maximum, with a mean squared error of 1.87. This result therefore implies that koligo is the same for the two variants, while the values of kd may differ slightly.
