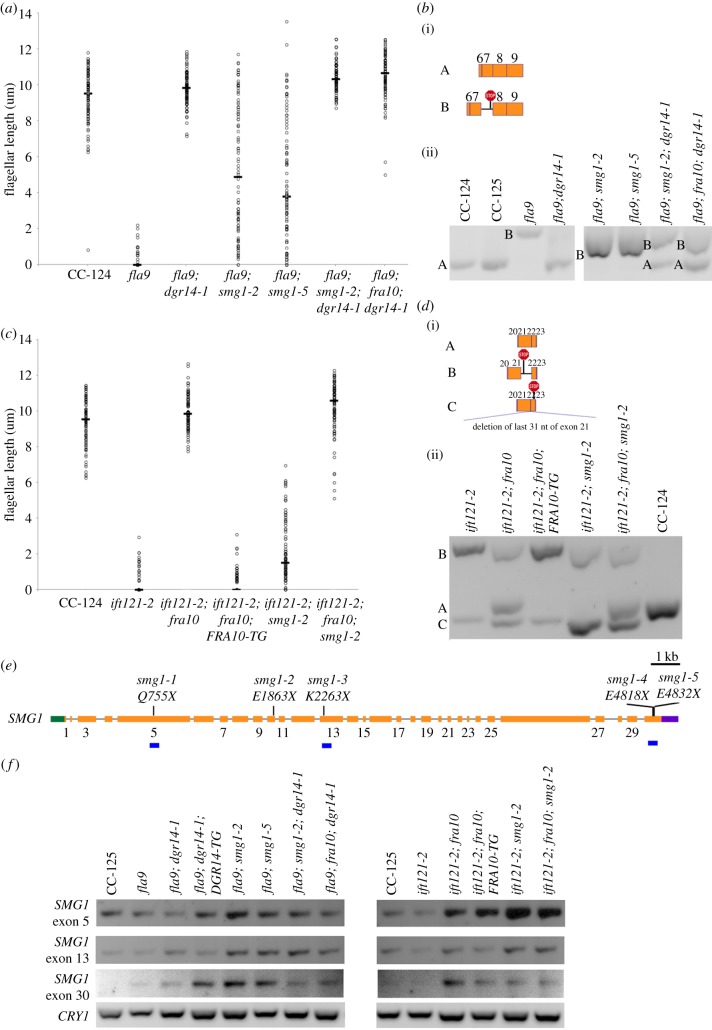
Figure 5.
Nonsense mutations in smg1 affect flagellar assembly in splice site ift mutants. (a) Distribution of flagellar length in wild-type (CC-124) and various fla9 mutants. Lengths of 100 flagella (represented by open circles) from 50 cells were measured in each strain. Horizontal bar represents the median flagellar length in each strain. (b) Alternative splicing of IFT81 between exons 6–9. (i) Representation of wild-type and intron retention transcripts. The red stop sign represents premature termination codon. (ii) RT-PCR products of IFT81 amplified from the same cells used in (a). (c) Distribution of flagellar length in wild-type (CC-124) and various ift121-2 mutants. Lengths of 100 flagella (represented by open circles) from 50 cells were measured in each strain. Horizontal bar represents the median flagellar length in each strain. (d) Alternative splicing of IFT121 between exons 20 to 23. (i) Representation of multiple IFT121 transcripts amplified within the region. (ii) RT-PCR products of IFT121 amplified from the same cells used in (c). (e) Gene structure of SMG1 and positions of multiple nonsense mutations. Green box, 5′ UTR; orange boxes, exons; black solid lines, introns; purple box, 3′ UTR; vertical black lines, positions of smg1 nonsense mutations. The positions of individual mutations in the SMG1 protein are indicated. Numbers of exons are indicated below the orange boxes. Owing to limitation of space, only odd numbers are included. The blue horizontal bars indicate regions amplified by RT-PCR in (f). (f) RT-PCR products of SMG1 in multiple exons. Amplification of the ribosomal protein gene CRY1 serves as a loading control.