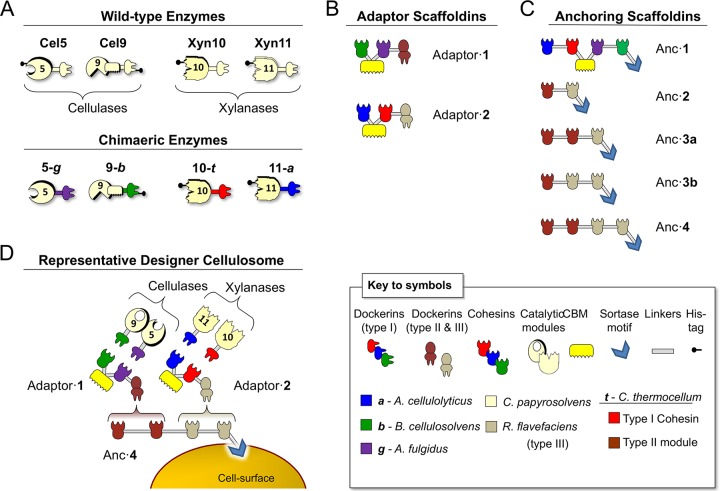
FIG 1.
Schematic representation of the wild-type and chimeric proteins used in this study. The bacterial species from which the representative modules are derived are shown in color-coded form in the pictograms. (A) Wild-type and chimeric C. papyrosolvens enzymes. In the shorthand notation for the recombinant enzymes, the numbers 5, 9, 10, and 11 correspond to members of the GH family of the respective catalytic modules; the origin of a given dockerin module is also indicated by lowercase italic characters as described in the key to the symbols. (B) Modular architectures of the two different adaptor scaffoldins designed for this work. Each example of adaptor scaffoldin contains two divergent cohesins for selective integration of different dockerin-containing enzymes and a type II or type III dockerin for attachment to the appropriate cohesin-containing anchoring scaffoldin. (C) Modular architectures of the various types of anchoring scaffoldins designed in this study. Each contains a C-terminal sortase signal motif for covalent attachment to the cell surface. Anc·1 contains 4 divergent cohesins for selective integration of 4 different dockerin-bearing enzymes. Anc·2 through Anc·4 are anchoring scaffoldins differing in numbers (2 to 4) or positions (3a or 3b) of cohesins that integrate the two adaptor scaffoldins and their resident enzymes. (D) Example of designer cellulosome assembly, resulting from a consortium of different strains of transformed L. plantarum.