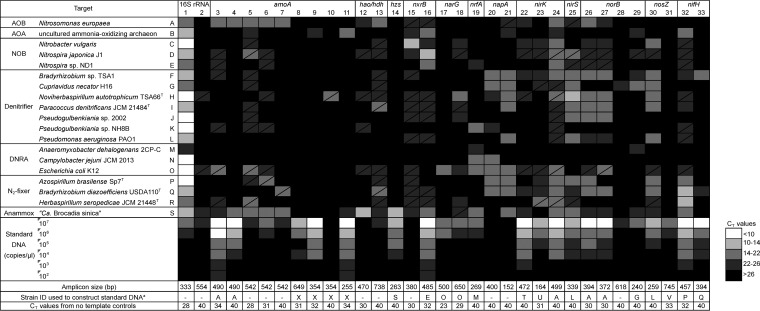
FIG 2.
Heat map showing the ranges in qPCR CT values on the NiCE chip for detection of N cycle functional genes and the 16S rRNA gene. The amplicon size, strain identification used to construct standard DNA, and CT values obtained from the nontemplate controls are shown at the bottom. The box with a diagonal line indicates the occurrences of nonspecific amplification. 1, primers 341F and 805R; 2, primers Archaea-F KO and Archaea-R KO; 3, primers amoA_1F and amoA_2R; 4, primers amoA_1F and amoA_F1_R2; 5, primers Gamo172 F1 and Gamo172 F1_R1; 6, primers Gamo172 F1 and Gamo172 F1_R2; 7, primers Gamo172 F2 and Gamo172 F2_R1; 8, primers Arch-amoAF and Arch-amoAR; 9, primers Arch-amoAFA and Arch-amoAR; 10, primers Arch-amoAFB and Arch-amoAR; 11, primers Arch-amoA-for and Arch-amoA-rev; 12, primers hzocl1F1 and hzocl1R2; 13, primers haoF4 and haoR2; 14, primers hzsA_1597F and hzsA1857R; 15, primers NxrB 1F and NxrB 1R; 16, primers nxrB169f and nxrB638r; 17, primers W9F and T38R; 18, primers narG1960f and narG2650r; 19, primers nrfAF2aw and nrfAR1; 20, primers V66 and V67; 21, primers V17m and napA4r; 22, primers FlaCu and R3Cu; 23, primers nirK876 and nirK1040; 24, primers nirK_166F and NxrB 1F; 25, primers nirSCd3aF and nirSR3cd; 26, primers norB2 and norB6; 27, primers cnorB-2F and cnorB-6R; 28, primers qnorB2F and qnorB7R; 29, primers qnorB2F and qnorB5R; 30, primers nosZ1F and nosZ1R; 31, primers nosZ-II-F and nosZ-II-R; 32, primers nifHF and nifHR; 33, primers IGK3 and DVV. a, strains used only to construct standard DNA: T, Pseudomonas sp. strain JCM 20650; U, Rhodobacter sphaeroides JCM 6121T; V, Gemmatimonas aurantiaca JCM 11422T; X, synthetic DNA fragment containing amoA from Nitrosopumilus maritimus SCM1 (GenBank accession no. CP000866; locus tag Nmar_1500).