Figure 1.
Mutation reference value to assure the quality of liquid biopsy–based results
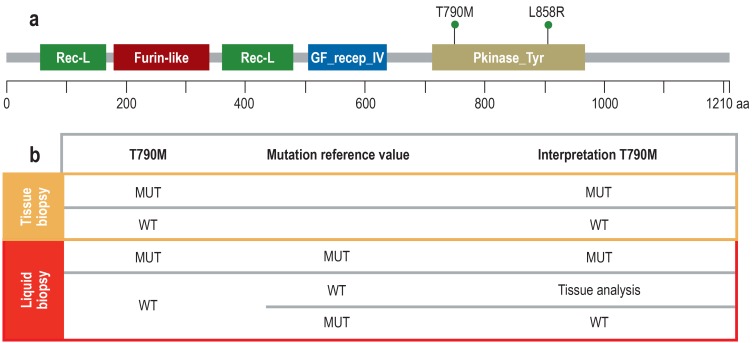
a) The diagram shows the epidermal growth factor receptor (EGFR) protein with domains (protein family [Pfam] nomenclature) and the two mutations L858R (activating mutation, exon 21) and T790M (resistance-mediated mutation, exon 20). The two mutations are located on the same deoxyribonucleic acids (DNA) strand (cis position) because the resistance-conferring T790M mutation only has an effect in the context of the activating mutation.
b) Since in tissue it is possible to exert histomorphological control over the number and proportion of tumor cells, the results of tumor genetic tests are reliable. In liquid biopsy, neither the absolute nor the relative proportion of ctDNA in the cfDNA is known; thus, any result is only reliable if a mutation (MUT) has been detected. If no mutation is detected, this can be due either to a wild type (WT) or to a sensitivity problem. However, in these cases it is helpful to analyze a mutation reference value. If this mutation can be detected, the amount of ctDNA present was adequate. Consequently, no T790M mutation (WT) is present. In the other case, a technical problem prevented mutation detection. Then, the tissue biopsy has to be analyzed to obtain a meaningful and clinically relevant test result. Since the T790M mutation represents a transition of cytosine (C) to thymine (T) (c.2369C>T), it can also occur by spontaneous deamination of C to uracil (U) which the mutation analysis would interpret as a mutation. Thus, it is important in this context too that the primary EGFR gene mutation is also tested for to be able to better distinguish between mutation and artificial transition.
Rec-L, receptor L-domain; Furin-like, furin-like cysteine rich region; GF recep_IV, growth factor receptor domain IV; Pkinase_Tyr, protein tyrosine kinase; aa, amino acid