Fig. 1. Comparative analysis of protein expressions between normoxic and hypoxic primed MSCs by SILAC-based quantitative proteomics.
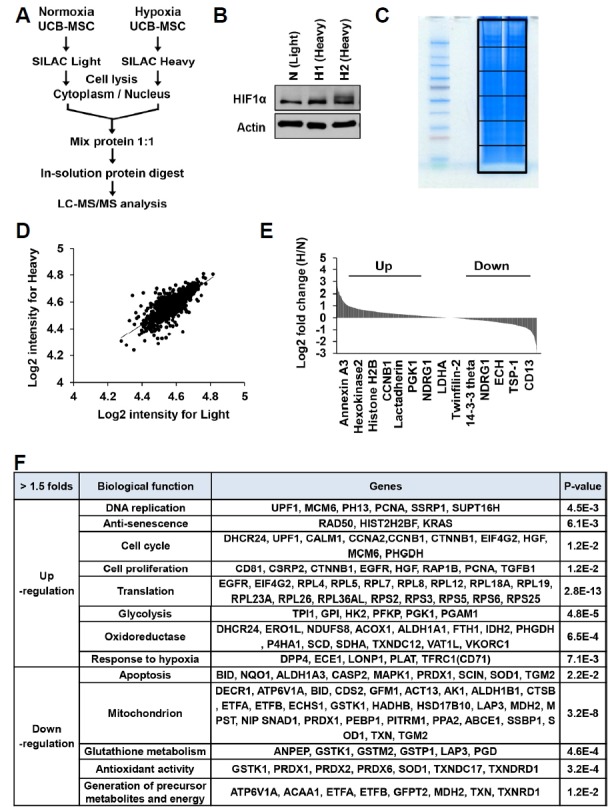
(A) Human umbilical cord blood-derived MSCs (MSCs) were cultured in normoxic (21% O2) or hypoxic (1% O2) conditions. Experimental scheme of SILAC labeling and LC-MS/MS analysis of normoxic and hypoxic primed MSCs at passage 3. (B) MSCs were grown in normoxic (light) and hypoxic (heavy) conditions, and analyzed by immunoblotting using anti-HIF1α antibody. (C) Both heavy and light samples were mixed, separated by SDS-PAGE, analyzed by in-gel-digestion, and then quantified by LC-MS/MS. (D, E) Ratio distribution of the 1361 differentially expressed proteins in hypoxic condition compared with the normoxic condition. Log2 SILAC ratios of 1.0 and −1.0 were the cut-offs for the regulated proteins (D). Representative upregulated (Up) and downregulated (Down) proteins by hypoxic conditioning were depicted (E). (F) List of proteins, which were upregulated or downregulated in hypoxic primed MSCs compared to the normoxic primed MSCs.
