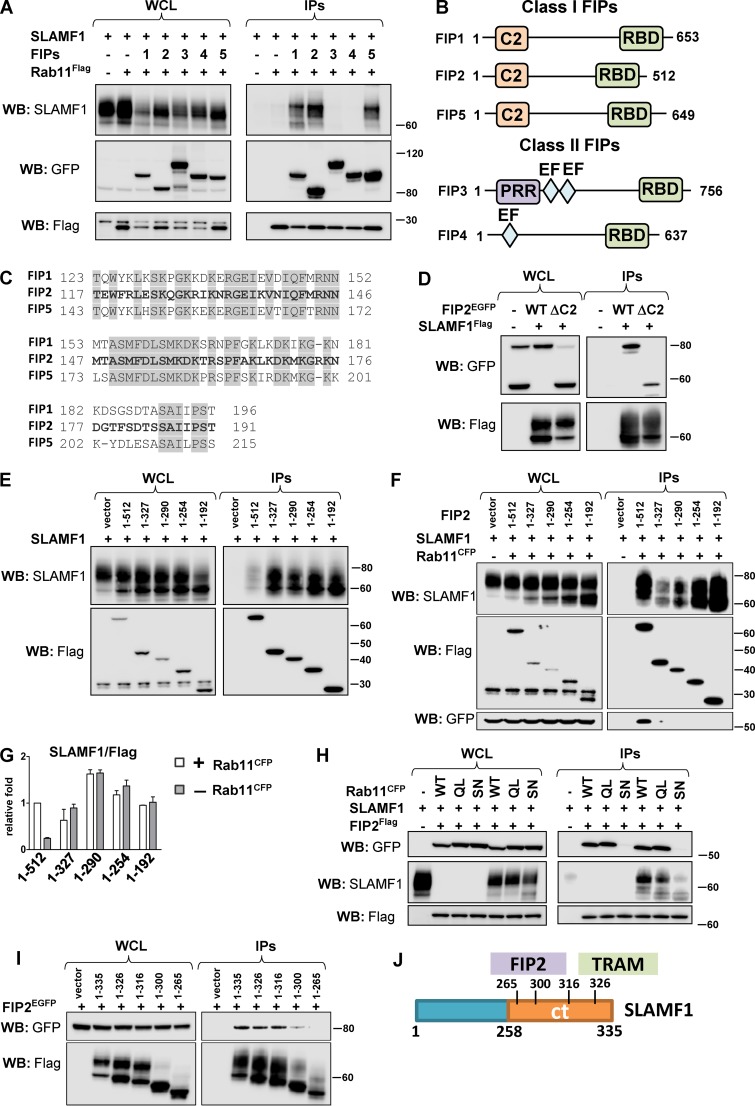
Figure 7.
SLAMF1 interacts with all class I Rab11 FIPs. (A) Anti-Flag IPs for Rab11aFlag with EGFP-tagged Rab11FIPs (1–5) and SLAMF1. (B) Schematic figure for class I and class II Rab11 FIPs domain structure. C2, phospholipid-binding C2 domain; EF, EF-hand domain; PRR, proline-rich region; RBD, Rab11 binding domain. (C) Homologous protein sequence in class I FIPs, which follow the C2 domain. Identical amino acids in all three class I FIPs are highlighted. (D) Coprecipitation of SLAMF1Flag with FIP2EGFP WT or FIP2 deletion mutant lacking the C2 domain (ΔC2). (E and F) Coprecipitation of untagged SLAMF1 with FIP2Flag (1–512 aa) and Flag-tagged FIP2 deletion mutants in anti-Flag IPs in the absence (E) or presence (F) of overexpressed Rab11CFP. (G) Quantification of coprecipitations in E and F between SLAMF1 and FIP2Flag variants correlated with the amount of Flag-tagged protein on the blot and Flag-tagged protein sizes. Error bars represent means ± SD for three independent experiments. (H) Coprecipitation of FIP2Flag with SLAMF1 and Rab11a WT, Rab11a Q70L mutant (QL), or Rab11a S25N mutant (SN). (I) Coprecipitation of SLAMF1Flag deletion mutants with FIP2EGFP. Molecular weight is given in kilodaltons. WB, Western blot; WCL, whole-cell lysate. (J) Scheme for FIP2- and TRAM-interacting domains in SLAMF1ct. The results are representative of at least three independent experiments.