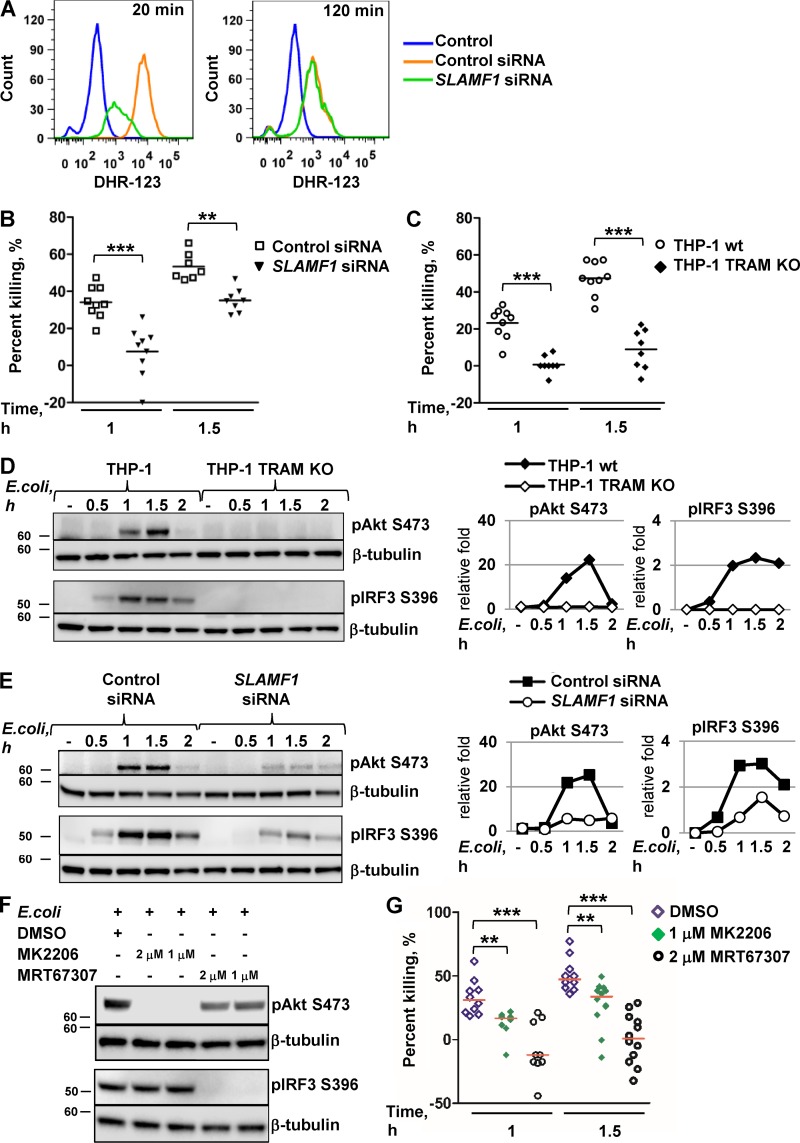
Figure 9.
TRAM and SLAMF1 are essential for the killing of E. coli by human macrophages. (A) Flow cytometry analysis of dihydrorhodamine 123 (DHR-123) fluorescence to access ROS activation in control siRNA or SLAMF1 siRNA human macrophages upon stimulation by E. coli red pHrodo particles. One of three experiments shown. (B and C) Bacterial killing assays by SLAMF1-silenced and control THP-1 cells (B) as well as TRAM KO and control THP-1 cells (C) infected with a DH5α strain at MOI 40. (D and E) Western blot analysis of pAkt (S473) and pIRF3 (S396) levels induced by E. coli particles in THP-1 WT and TRAM KO cells (D) as well as SLAMF1-silenced or control oligonucleotide–treated cells (E). Graphs (right) on Western blotting show quantification of protein levels relative to β-tubulin obtained with Odyssey software. (F) Western blot showing phospho-(S396) IRF3 and phospho-(S473) Akt levels in lysates of THP-1 cells coincubated with E. coli particles for 1 h in the presence or absence of TBK1-IKKε inhibitor (MRT67307), pan-Akt allosteric inhibitor (MK2206), or DMSO. Molecular weight is given in kilodaltons. (G) Bacterial killing assays by THP-1 cells with DMSO (<0.01%), 1 μM Akt inhibitor MK2206, or 2 μM TBK1-IKKε inhibitor MRT67307 upon infection by DH5α at MOI 40. Percent killing was calculated as 100 − (number of CFUs at time X/number of CFUs at time 0) × 100 for average values of technical replicates, and each dot on the graphs in B, C, and G represents a biological replicate from three independent experiments. Median values are shown by lines. Statistical significance was calculated by a Mann-Whitney nonparametric test. **, P < 0.01; ***, P < 0.001.