Abstract
In cancer cells, the glycoprotein Mucin 1 (MUC1) undergoes abnormal, truncated glycosylation. The truncated glycosylation exposes cryptic peptide epitopes that can be recognized by antibodies. Since these immunogenic regions are cancer specific, they represent ideal targets for therapeutic antibodies. We investigated the role of tumor-specific glycosylation on antigen recognition by the therapeutic antibody AR20.5. We explored the affinity of AR20.5 to a synthetic cancer-specific MUC1 glycopeptide and peptide. The antibody bound to the glycopeptide with an order of magnitude stronger affinity than the naked peptide. Given these results, we postulated that AR20.5 must specifically bind the carbohydrate as well as the peptide. Using X-ray crystallography, we examined this hypothesis by determining the structure of AR20.5 in complex with both peptide and glycopeptide. Surprisingly, the structure revealed that the carbohydrate did not form any specific polar contacts with the antibody. The high affinity of AR20.5 for the glycopeptide and the lack of specific binding contacts support a hypothesis that glycosylation of MUC1 stabilizes an extended bioactive conformation of the peptide recognized by the antibody. Since high affinity binding of AR20.5 to the MUC1 glycopeptide may not driven by specific antibody–antigen contacts, but rather evidence suggests that glycosylation alters the conformational equilibrium of the antigen, which allows the antibody to select the correct conformation. This study suggests a novel mechanism of antibody–antigen interaction and also suggests that glycosylation of MUC1 is important for the generation of high affinity therapeutic antibodies.
Keywords: cancer, crystal structure, immunotherapy, molecular recognition, mucin
Introduction
Mucin 1 (MUC1; also known as polymorphic epithelial mucin (PEM), CA15-3, EMA, MCA and episialin) is a membrane glycoprotein that has been identified as an important target for cancer immunotherapy (Kimura and Finn 2013). MUC1 is a type I transmembrane heterodimer, comprised of two subunits: an α-subunit consisting of an extracellular N-terminal domain and a β-subunit composed of a transmembrane helix and cytoplasmic tail. The extracellular domain of MUC1 contains a variable number of tandem repeats (VNTR) consisting of 20–120 repeats of a 20-amino acid sequence (HGVTSAPDTRPAPGSTAPPA). Within the VNTR sequence, there are five potential O-glycosylation sites (underlined above). In healthy tissues, the protein is heavily O-glycosylated, with 50–90% of the protein weight being carbohydrate (Apostolopoulos et al. 2015). During neoplastic transformation, MUC1 is overexpressed and as a result of altered glycosyltransferase activity it undergoes incomplete glycosylation. The appearance of short, truncated carbohydrate side chains including the Tn antigen (GalNAc), TF antigen (Gal(β1-3)GalNAc) and STn or the sialyl Tn antigen (Neu5Ac(α2-6)GalNAc), exposes MUC1 as a tumor antigen. All five glycosylation sites in the VNTR repeat sequence can be glycosylated in tumor cells (Muller et al. 1999). MUC1 is overexpressed and altered in over half of cancer cases, and as such is an attractive target for immunotherapy (Acres and Limacher 2005). A variety of therapeutic antibodies and synthetic peptide/glycopeptide vaccines have been developed (Acres and Limacher 2005; Kimura and Finn 2013). Despite the wealth of clinical and immunological data available on MUC1 antibodies, there is surprisingly little known regarding the molecular details by which MUC1-specific antibodies recognize their targets. To date, the only X-ray structures available of a MUC1-specific antibody in complex with antigen is that of mAb SM3 in complex with both unglycosylated and glycosylated MUC1 (Dokurno et al. 1998; Martinez-Saez et al. 2015).
mAb AR20.5 (OncoQuest Inc., Edmonton, AB, Canada) is a murine anti-MUC1 monoclonal antibody (IgG1) produced by immunization with MUC1 from an ovarian cancer patient (Qi et al. 2001). The antibody is under investigation for therapeutic potential, having undergone a successful phase I clinical trail with no observed toxicity (de Bono et al. 2004). Unlike traditional therapeutic antibodies that directly target and either destroy the tumor via immune mechanisms or pharmacologically modified receptor biology (Nicodemus 2015), AR20.5 is thought to function in a vaccine-like manner by generating a tumor-associated MUC1-specific immune response in the patient. Low dose delivery of the antibody results in binding of serum-circulating MUC1. The antibody–antigen immune complex is internalized by dendritic cells, which in turn activate cytotoxic T cells. The activated T cells infiltrate and eradicate the tumor. In support of this model of AR20.5 action, dendritic cells pulsed ex vivo with MUC1–AR20.5 complexes induced CD4+ and CD8+ T cells (Schultes et al. 2005).
AR20.5 binds the MUC1 VNTR consisting of the immunodominant DTR portion of the antigen. Pepscan epitope mapping revealed that the core epitope recognized by AR20.5 consists of six amino acids within VNTR region (DTRPAP). Furthermore, low affinity was reported after inhibiting MUC1 glycosylation in vivo, suggesting that the carbohydrate moiety could be involved in antigen recognition and binding by mAb AR20.5 (Qi et al. 2001).
The involvement of carbohydrate residue epitopes in antibody binding has been studied for several MUC1 mAbs (Hilgers 1998; Weiner 2007). Antigen glycosylation was initially regarded as a hurdle for high affinity binding by therapeutic antibodies, as it was believed that the immunodominant DTR sequence was not glycosylated in cancer cells (Finn et al. 1995). However, this view has been challenged as accumulating evidence points to the importance of tumor-associated carbohydrates in MUC1 antibody binding, and for the generation of a robust immune response in vaccine candidates. A comprehensive binding study on a panel of MUC1 antibodies concluded that all mAbs generated by immunization from tumor MUC1 displayed increased affinity for glycosylated MUC1 compared to the unglycosylated antigen. Whereas, antibodies generated by immunization with synthetic peptide or nontumor-derived MUC1 did not exhibit higher affinity binding for the glycosylated form of MUC1 (Hilgers 1998; Karsten et al. 1998). Similar results have been found in MUC1 vaccine studies, where vaccine strategies incorporating unglycosylated MUC1 fragments have been largely unsuccessful, whereas the incorporation of MUC1 glycopeptide vaccine candidates has shown therapeutic promise (Lakshminarayanan et al. 2012; Abdel-Aal et al. 2014; Glaffig et al. 2015; Thompson et al. 2015).
While the importance of tumor-associated MUC1 glycosylation in antibody binding is clear, key questions remain regarding the molecular mechanism behind how the carbohydrate induces higher affinity binding. There are a variety of plausible mechanisms by which antigen glycosylation could influence antibody binding. The carbohydrate could directly form part of the epitope, as observed in the glycopeptide-specific antibody 237 mAb (Brooks et al. 2010), the carbohydrate could form specific, but nonessential contacts with the antibody which influences affinity as observed in the MUC1 mAb SM3 (Martinez-Saez et al. 2015), or the carbohydrate could influence the structure of the antigen. A detailed understanding of how AR20.5 recognizes MUC1 and the elucidation of the molecular mechanisms by which glycosylation modulates antigen binding will provide valuable information within the context of MUC1 cancer vaccine design and future development of MUC1 therapeutic antibodies.
Results
Sequencing and germline analysis of AR20.5
Multiple light chain clones (12) and heavy chain clones (6) were sequenced: 12 of the 14 light chain clones produced identical nucleotide sequences, while the other 2 sequences were similar to each other, but distinct from the other 12. Surprisingly, examination of the sequences using the IMGT database to check for productive VDJ gene rearrangements (Lefranc et al. 2009; Lefranc 2014) revealed that the 12 identical light chain nucleotide sequences were pseudogenes that contained a stop codon in the coding sequence and a frameshift mutation at the junction. The presence of pseudogenes and multiple transcripts from hybridomas is unusual, but has been known to occur (Carroll et al. 1988). Analysis of the remaining 2 light chain sequences indicated productive gene rearrangements, and thus it was concluded that these sequences represented true antibody sequence. All the heavy chain sequences were nearly identical, and appeared to be the result of a productive gene arrangement.
Germline gene analysis using IMGT V-quest suggested that the light chain originated from V-gene IGKV1-117*01 (97.3% identical) and J-gene IGKJ1*01 (100% identical). The heavy chain likely originated from V-gene, IGH5-12-2*01 (97.2% identical), J-gene IGHJ2*01 (93.8% identical) and D-gene IGHD1-1*01. Alignment of the translated germline sequences with AR20.5 revealed sites of putative somatic hypermutation. The light chain had a total of four mutations, three in complementarity determining region (CDR) L1 (S28T, N35K, T36I) and a mutation in framework region 3 (N58K). The heavy chain had a total of six putative amino acid substitutions from the germline sequence. There was a single mutation in CDR H1 (T33P), three mutations in CDR H2 (S52N, S57N, T58P) and two mutations in framework region 3 (M93I, A97I).
Glycosylation of a MUC1 peptide with a Tn antigen enhances binding to AR20.5
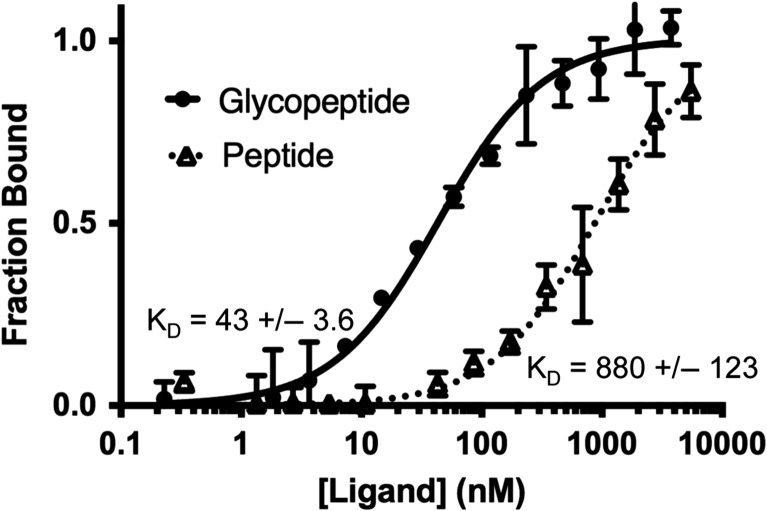
AR20.5 binds a 6-amino acid epitope (DTRPAP) within the repeating segment of 20 amino acids in the MUC1 VNTR domain (Qi et al. 2001). The AR20.5 epitope contains a single putative glycosylation site at Thr. To determine the impact of epitope glycosylation on AR20.5 affinity for MUC1, an 8-MER synthetic peptide (APDTRPAP) and corresponding glycopeptide with a single Tn antigen (Thr-GalNAc) on the Thr glycosylation site (APDTnRPAP) was synthesized. Microscale thermophoresis (MST) was used to determine the affinity of AR20.5 Fab for both peptide and glycopeptide. During MST experiments, a clear concentration-dependent enhancement of fluorescence signal was observed. In cases where a significant change in fluorescence occurs, the MST phenomenon cannot be used for KD determination. To confirm that the observed change in fluorescence was caused by the binding event, a denaturation test was carried out which resulted in an abrogation of the enhanced fluorescence signal. The results of the denaturation test indicate that the binding event was responsible for the observed signal change in fluorescent intensity. The change in fluorescent signal was then used to determine KD (Figure 1). AR20.5 bound to the MUC1 peptide with a KD of 880 ± 123 nm. The high error in the KD determination is the result of incomplete binding saturation within the solubility range of the peptide. The glycopeptide bound to the antibody with a KD of 43 ± 3.6 nm. Addition of a single GalNAc residue enhances the binding of AR20.5 for antigen by a factor of ~20.
Fig. 1.
Binding of AR20.5 Fab to a MUC1 peptide and glycopeptide. Binding affinity of AR20.5 Fab fragment to synthetic MUC1 peptide (APDTRPAP) (dashed line, triangles) and glycopeptide (APDTnRPAP) (solid line, closed circles) was determined by monitoring the ligand-binding induced change in the fluorescence of labeled AR20.5. Error bars represent the standard error of the mean from three separate experiments. MUC1 glycopeptide binds to AR20.5 with ~20-fold higher affinity compared to the peptide.
Overview of the structures of AR20.5 co-crystallized with MUC1 peptide and glycopepitde
To ascertain the structural basis for the difference in affinity of AR20.5 for glycosylated vs. nonglycosylated peptide epitope, AR20.5 Fab was co-crystallized with the synthetic MUC1 peptide/glycopeptide corresponding to the epitope region (APDTRPAP and ADPTnRPAP). The protein crystallized in different space groups for the two ligands despite identical conditions for co-crystallization (Table I). Both structures contain two Fab molecules in the asymmetric unit and are arranged in a head-to-tail fashion. The conformation of the Fab polypeptide backbone was nearly identical in both peptide and glycopeptide complexed structures (0.21 Å2). The Fab structures exhibited a disordered region within a solvent exposed loop of the heavy chain constant domain (GlyH130–ThrH135).
Table I.
Data collection and refinement statistics
| Peptide | Glycopeptide | |
|---|---|---|
| Data processing | ||
| Space group | P 31 21 | C 2 2 21 |
| Cell parameters (Å) | 69.7, 69.7, 363.43, 90, 90, 120 | 97.24, 100.45, 235.29, 90, 90, 90 |
| Resolution (Å) | 46.44–1.91 (2.04–1.97)a | 48.62–2.20 (2.28–2.20) |
| Total reflections | 7,43,710 (76,637) | 432,636 (43,680) |
| Unique reflections | 74,059 (7301) | 58,698 (5765) |
| Completeness (%) | 99.8 (100) | 100.00 (100.00) |
| Multiplicity | 10 (10.5) | 7.3 (7.6) |
| Rmerge (%) | 11.27 (67.08) | 8.81 (125.5) |
| CC1/2 (%) | 99.6 (0.812) | 99.8 (70.3) |
| <I/σI > | 12.19 (3.61) | 12.18 (1.48) |
| Wilson B (Å2) | 32.84 | 50.87 |
| Refinement | ||
| Rwork/Rfree (%) | 21.89/23.42 | 19.41/22.11 |
| RMS | ||
| Bonds (Å) | 0.003 | 0.009 |
| Angles (°) | 0.82 | 1.03 |
| Average B (Å2) | ||
| Protein | 44.30 | 64.40 |
| Solvent | 41.90 | 59.81 |
| Ligand | N/A | 61.23 |
| Molprobity validation | ||
| Ramachandran plot | ||
| Most favored (%) | 98 | 97 |
| Additionally allowed (%) | 1.6 | 2.4 |
| Disallowed (%) | 0.24 | 0.24 |
| Rotamer outliers (%) | 0 | 0 |
| Clashscore | 1.07 | 1.90 |
| PDB code | 5T6P | 5T78 |
aValues given in parentheses correspond to the outer shell of data.
Structure of AR20.5 with MUC1 peptide
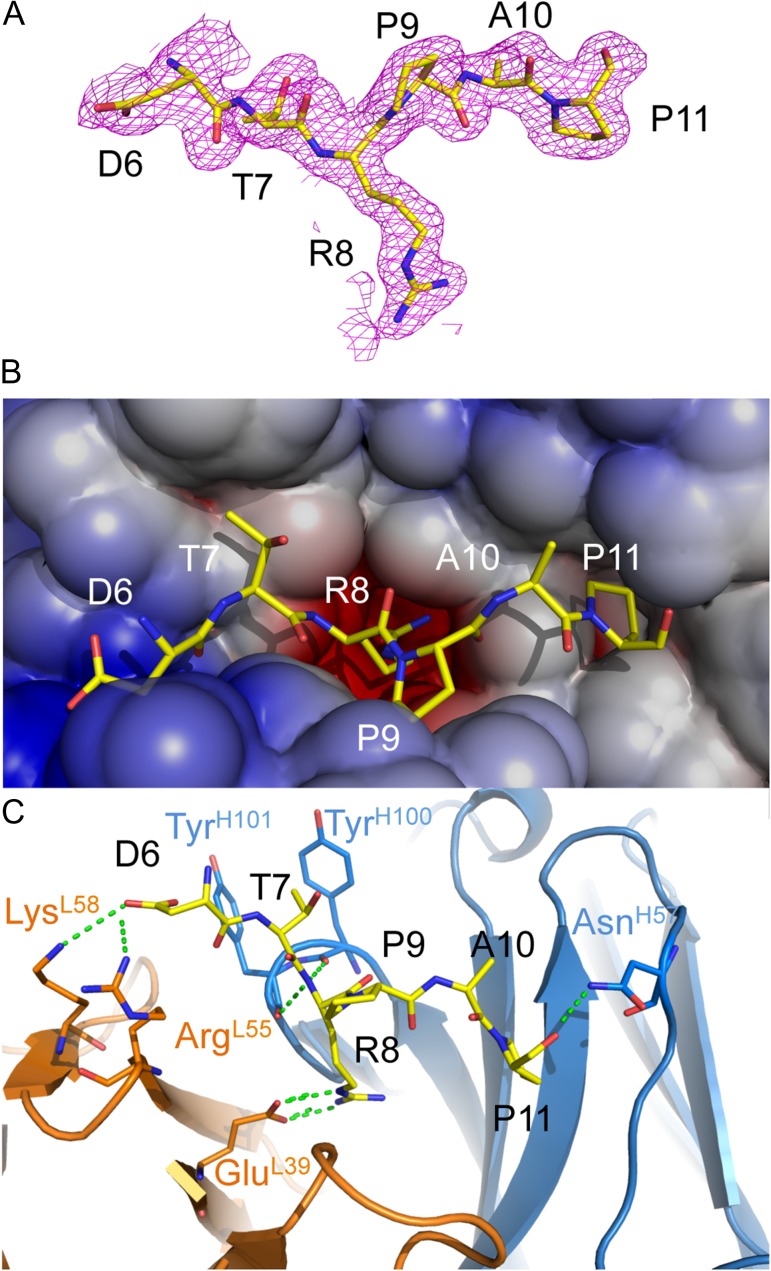
In the structure of AR20.5 in complex with a synthetic MUC1 peptide (APDTRPAP), the region corresponding to the known AR20.5 epitope (DTRPAP) exhibited clear unambiguous electron density in 2Fo-Fc maps following molecular replacement and an initial cycle of refinement (Figure 2A). The first two N-terminal amino acids from the synthetic ligand (Ala and Pro) were disordered, suggesting that these residues are not critical for recognition. The AR20.5 combining site forms a surface groove and pocket ringed by CDRs L1, L2, H2 and H3. In the structure, the central Arg residue in the MUC1 epitope (R8) acts as an anchor, binding into a deep hydrophobic and electronegative pocket walled by residues on both the light chain (PheL94, TrpL101, GluL39, TyrL37) and the heavy chain (GlyH102, PheH103, AsnH99, TyrH50). The rest of the epitope lies in a surface groove (Figure 2B). The binding of the peptide results in a significant amount of the ligand surface area buried from solvent. Upon complex formation, a total of 728 Å2 of the MUC1 peptide is buried, corresponding to 78% of the total solvent accessible surface area. Remarkably, the MUC1 VNTR peptide epitope makes only a few polar contacts with the antibody-combining site. The light chain forms a total of three salt-bridges to the epitope (GluL39 to R8, ArgL55 and LysL58 to D6) and the heavy chain forms three hydrogen bonds to the ligand (TyrH100 to R8, TyrH101 to T8 and AsnH57 to P11) (Figure 2C).
Fig. 2.
X-ray structure of AR20.5 in complex with MUC1 peptide (APDTRPAP). (A) 2Fo-Fc electron density map of MUC1 peptide bound to AR20.5. The N-terminal residues A4 and P4 were disordered in the structure. (B) Electrostatic surface of AR20.5 combining site. The MUC1 peptide antigen binds in a surface groove. Residue R8 of MUC1 binds in a deep negatively charged pocket. (C) Binding interactions of MUC1 peptide (yellow) with AR20.5. Binding is mediated by a series of electrostatic interactions and hydrogen bonds to both the heavy chain (blue) and the light chain (orange). This figure is available in black and white in print and in color at Glycobiology online.
Structure of AR20.5 with Tn-MUC1 glycopeptide
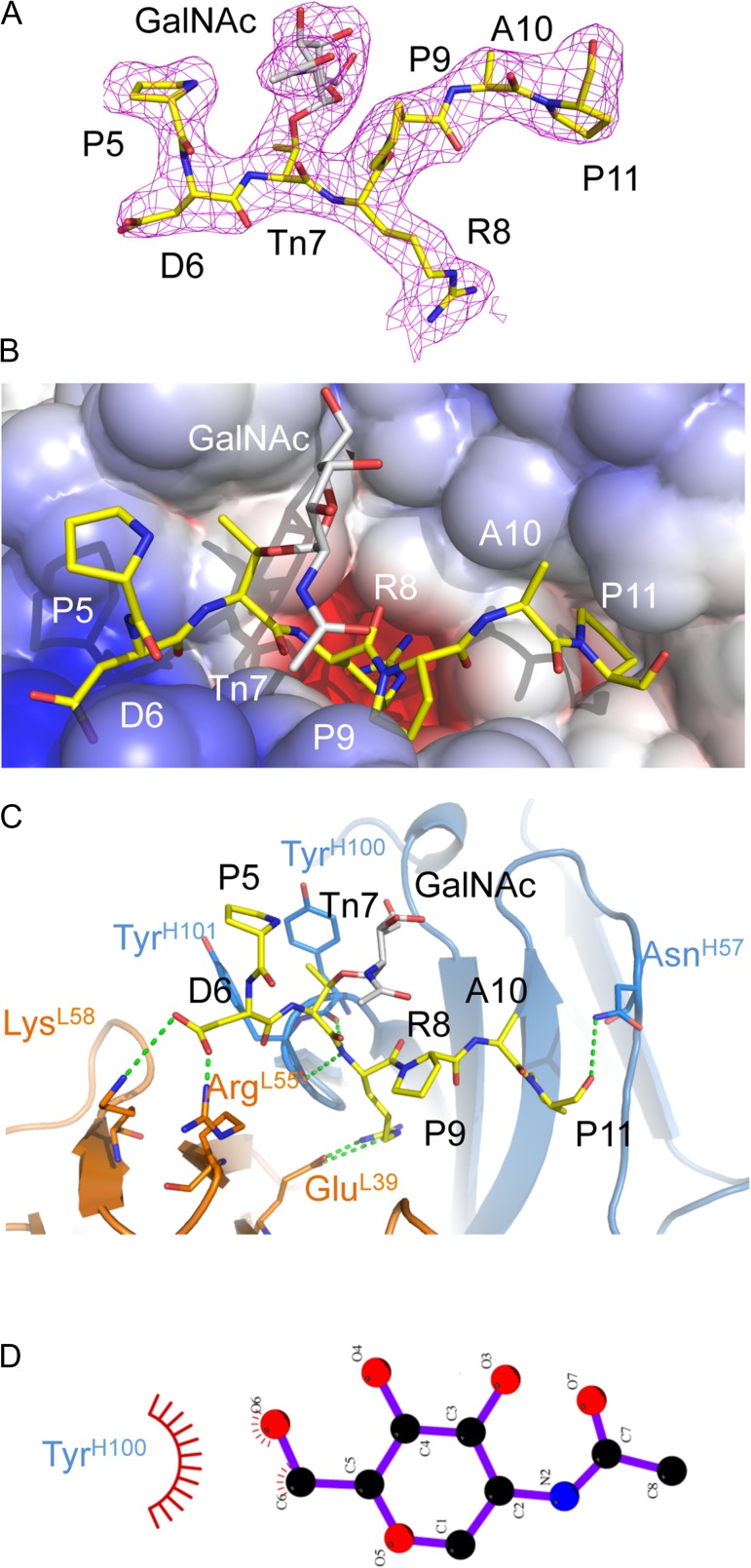
The structure of AR20.5 in complex with a GalNAc glycosylated (Tn antigen) glycopepitde (APDTnRPAP) revealed clear unambiguous electron density in 2Fo-Fc maps following molecular replacement and an initial cycle of refinement for the region corresponding to the known AR20.5 epitope (DTRPAP) (Figure 3A). Unlike the peptide structure, only the first N-terminal amino acid (Ala) was disordered, with the Pro (P5) residue being visible, albeit with a high B-factor, suggesting a high degree of mobility.
Fig. 3.
X-ray structure of AR20.5 in complex with MUC1 glycopeptide (APDTnRPAP). (A) 2Fo-Fc electron density map of MUC1 glycopeptide bound to AR20.5. The N-terminal residue A4 was disordered in the structure. There is clear electron density for the GalNac carbohydrate (gray). (B) Electrostatic surface of AR20.5 combining site. The MUC1 glycopeptide antigen binds in a surface groove. Residue R8 of MUC1 binds in a deep negatively charged pocket. (C) Binding interactions of MUC1 glycopeptide (yellow) with AR20.5. Binding is mediated by a series of electrostatic interactions and hydrogen bonds to both the heavy chain (blue) and the light chain (orange). The GalNAc carbohydrate (gray) makes no specific polar contacts with the antibody. (D) Nonpolar interactions between GalNAc and Tyr100H figure generated with LigPlot+ (Laskowski and Swindells 2011). This figure is available in black and white in print and in color at Glycobiology online.
The binding interactions of AR20.5 with the glycopeptide were essentially identical to those observed in the peptide structure. The glycopeptide lies in an identical position in the AR20.5 combining site, with R8 being buried in a pocket, and the rest of the glycopeptide lying in a surface groove (Figure 3B). The GalNAc residue does not point into the combining site, but rather faces away from the binding site toward the solvent (Figure 3B). There is a slight increase in the total surface area being buried from solvent upon complex formation with the glycopeptide, compared to the peptide, with a total of 820 Å2 being buried. The increase in buried surface is attributed to the carbohydrate (GalNAc) with 31% of the carbohydrate solvent accessible surface area being buried by complex formation.
Interestingly despite the addition of the GalNAC residue, the binding interactions to the glycopeptide are identical to that of the peptide. The same set of salt-bridges and hydrogen bonds is formed on the peptide portion of the MUC1 glycopeptide. The only visible difference between the observed interactions was an additional H-bond visible to the partially ordered P5 residue. The GalNAc carbohydrate points itself that it does not appear to directly participate in the binding interaction, as it makes no specific polar contacts to the antibody-combining site (Figure 3C).
Circular dichroism spectroscopy of synthetic MUC1 peptide and Tn glycopeptide
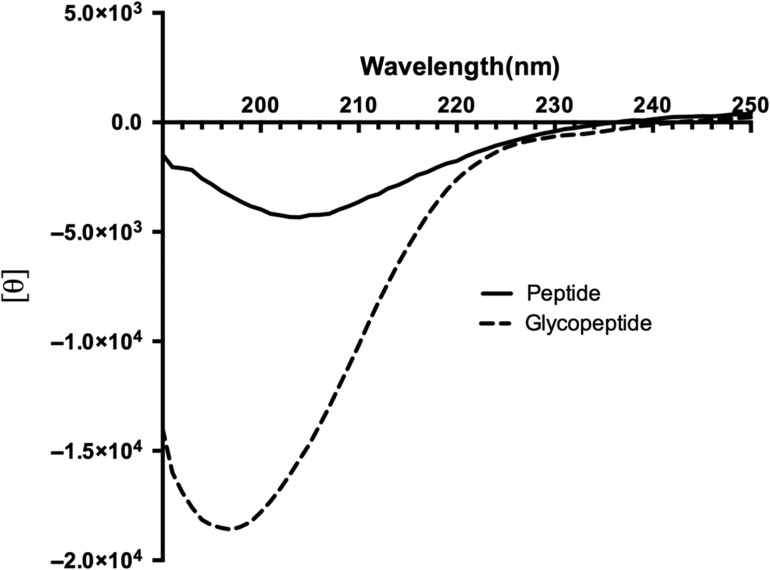
The solution conformation of the 8-MER MUC1 peptide and Tn glycopeptide (APDTRPAP and ADPTnRPAP) was examined by recording the circular dichroism (CD) spectrum from 290 to 190 nm (Figure 4). As expected for such a short fragment, neither peptide displayed a spectra characteristic of well-ordered secondary structures. The unglycsolyated MUC1 peptide displayed a strong negative peak at 205 nm (Figure 4), which is typical of a type I β-turn (Greenfield 2006). The Tn glycopeptide exhibited a larger negative peak owing to the presence of carbohydrate (Gururaja et al. 1998; Uray et al. 2014), furthermore the peak maxima were shifted to 196 nm which is the characteristic of a disordered structure (Greenfield 2006) (Figure 4).
Fig. 4.
Circular dichroism (CD) spectrum of 8-MER MUC1 peptide (APDTRPAP) and glycopeptide (ADPTnRPAP). The MUC1 peptide displayed a strong negative peak at 205 nm typical of a type I β-turn. The glycopeptide exhibited a larger negative peak owing due to the carbohydrate with the peak maxima shifted to 196 nm which is the characteristic of a disordered structure.
Discussion
Tumor-associated glycosylation influences MUC1 antibody binding
The addition of a single GalNAc (Tn antigen) residue on the Thr residue of the DTRPAP sequence dramatically enhanced the binding of AR20.5 to the MUC1 epitope (Figure 1). This conclusion is further supported by the previous observation that deglycosylation of surface expressed MUC1 on OVCAR-3 cells decreased AR20.5 binding (Qi et al. 2001). Over the past several decades, numerous MUC1-specific antibodies have been generated for both research and clinical applications. Many of these antibodies bind the immunodominant DTR epitope of MUC1 (Burchell et al. 1987). While it was initially believed that the fully deglycosylated form of DTR represented the tumor-specific epitope (Finn et al. 1995), later it was discovered that the DTR sequence is glycosylated with short glycans, including the Tn antigen (Muller et al. 1997). Indeed, a common feature of many (but not all) MUC1 antibodies appears to be the ability to bind the naked (aglycone) form of the MUC1 peptide, but at the same time exhibiting higher affinity binding to glycosylated forms of the antigen (Karsten et al. 1998; Möller et al. 2002; Matsushita et al. 2014). Interestingly, a recent study evaluating the involvement of cancer-specific glycans on MUC1 antibody reactivity came to the conclusion that DTR-specific antibodies generated by immunization from tumor sources displayed a higher affinity for glycosylated VNTR peptides than unglycosylated peptides, suggesting that the tumor-specific immunodominant epitope is actually a hypoglycosylated form of MUC1 (Karsten et al. 2004). In further support of the hypoglycosylated form, rather than a completely deglycosylated form of MUC1 being the tumor-specific epitope, the observation is that glyco-specific MUC1 antibodies have been associated with improved prognosis in breast cancer patients (Blixt et al. 2011). Tumor-specific carbohydrates on MUC1 may also be important for activation of the cellular immune response as it has been reported that glycans are not removed during antigen processing (Vlad et al. 2002) and the GalNAc moiety was detected during the antigen presentation to the T cells (Apostolopoulos et al. 2003). Such evidence strongly suggests that the native epitope of a tumor-related MUC1 is glycosylated. Clearly, tumor-specific glycosylation of MUC1 is important in antigen recognition by MUC1-specific antibodies. And may explain for the activity of AR20.5 as an inducer of anti-MUC1-associated tumor immunity in patients; however, question remains: what is the molecular basis for the observed higher affinity binding?
Molecular basis for the high affinity binding of AR20.5 to Tn-glycosylated MUC1?
There are three plausible explanations for the observed higher affinity binding of AR20.5 and other antibodies to glycosylated forms of MUC1: (i) the glycan forms a part of the epitope and the antibody binds to the sugar; (ii) the carbohydrate can form specific but nonessential contacts with the antibody which influences affinity, (iii) glycosylation influences the conformation of the epitope recognized by the antibody in some manner which affects the affinity of the antibody for the antigen.
Unlike mAb SM3, AR20.5 does not directly bind the glycan in the DTnR epitope
It has been previously demonstrated the glycan in a glycopeptide can directly form a part of the epitope recognized by a monoclonal antibody. The first high-resolution X-ray structure of an antibody in complex with a glycopeptide demonstrated that it is possible for both the carbohydrate and the peptide portion of a glycopeptide antigen to make specific contacts with the antibody and thus form the epitope (Brooks et al. 2010). The antibody 237 mAb in complex with a Tn glycopeptide derived from the tumor-associated protein Podoplanin revealed that the GalNAc residue acted as an anchor, and was buried in a pocket and was heavily hydrogen bonded, while the peptide portion of the antibody bound to a surface groove (Brooks et al. 2010). Interestingly, the MUC1-specific antibody SM3 exhibits similar behavior to AR20.5, as the addition of GalNAc to the DTR regions of MUC1 resulted in a 3-fold increase in affinity. However, the X-ray structure of SM3 in complex with a MUC1 glycopeptide suggested that the observed higher affinity to the Tn-glycosylated peptide was mediated by a hydrogen bond to the side chain of Tyr32L and stacking of the acetyl group to the side chain of Trp33H (Martinez-Saez et al. 2015). However, despite two antibody X-ray structures that detail some specific interaction of the glycan with the antibody, this mechanism of recognition observed in SM3 may not be occurring in AR20.5. The GalNAc residue in the Tn antigen does not appear to directly participate in binding, as no specific hydrogen bonds or stacking interactions are formed between the sugar and the antibody-combining site (Figure 3C). The only possible interactions between GalNAc and the antibody-binding site are a potential weak hydrogen bond and van der Waal contact with Tyr100H. However, the hydroxymethyl of GalNAc is positioned 4 Å away from the side chain of Tyr100H and is unlikely to form a energetically significant hydrogen bond (Jeffrey 1997). While the carbohydrate forms van der Waal interaction with Tyr100H (Figure 3D), these interactions may contribute slightly to enhanced binding affinity to the carbohydrate; however, they are unlikely sufficient to explain the 20-fold increase in affinity compared to the nonglycosylated peptide.
Cancer-specific epitope recognized by AR20.5 is and a extended conformation
Early work examining the conformation of VNTR repeats determined that in the absence of glycosylation, the immunodominant PDTR sequence adopted a β-turn structure (type I) (Scanlon et al. 1992; Fontenot et al. 1993, 1995; Liu et al. 1995). Since nearly all MUC1 antibodies are capable of binding the naked, unglycosylated VNTR, it was initially presumed that the type I β-turn structure represented the conformational epitope recognized by (P)DTR-specific MUC1 antibodies. Given the observation that the binding of MUC1 antibodies has enhanced glycosylation, it was logical to hypothesize that the glycan either formed part of the epitope or stabilized the β-turn epitope.
A β-turn is composed of four consecutive residues where the distance between Cαi and Cαi+3 is less than 7 Å and the turn results in a change in the direction of the protein chain. β-Turns are also frequently accompanied by specific hydrogen bonding patterns (NHi+3–COi) and approximate dihedral angles (Φ2 = −60°, ψ2 = −30°; Φ3 = −90°, ψ3 = 0°) (Richardson 1981).
The results of the CD experiments on the 8-amino acid MUC1 peptide used in this study confirmed that the predominant form of the MUC1 PDTR region is a type I β-turn (Figure 4). However, AR20.5 clearly binds an extended conformation of the peptide as there is no evidence of a type I β-turn in the structure (Table II, Figure 5A). If the PDTR formed a type I β-turn, Pro would be less than 7 Å from Arg, and Asp would have phi/psi angles of −60°/−30°, and Thr would have phi/psi angles of ~ −0°, 0°.
Table II.
Phi/Psi angles of the PDTR region of MUC1 determined by X-ray crystallography and NMR
| MUC1 peptide | MUC1 glycopeptide | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residue | This studya | Dokurno et al. (1998)a | Martinez-Saez et al. (2015)a | Schuman et al. (2003)b | This studya | Martinez-Saez et al. (2015)a | ||||||||
| Φ | Ψ | Φ | Ψ | Φ | Ψ | Φ | Ψ | Φ | Ψ | Φ | Ψ | Φ | Ψ | |
| A | −92.3 | 160.0 | −65.3 (31.8) | 145.9 (38.0) | −66.3 (31.8) | 145.8 (38.0) | ||||||||
| P | −80.5 | −164.6 | −81.4 | −157.5 | −67.2 (42.4) | 141.6 (49.9) | −78.9 | −157.4 | −64.3 (31.8) | 142.2 (38.0) | ||||
| D | −78.5 | 100.6 | −76.0 | 102.7 | −66.2 (31.8) | −44.4 (38.0) | −100.1 | 26.4 | −77.5 | 88.5 | −81.8 (31.8) | 148.8 (38.0) | ||
| T/Tn | −76.1 | 154.7 | −97.2 | 16.0 | −94.7 | 8.7 | −86.3 (42.4) | −15.9 (49.9) | −69.4 | 148.1 | −87.8 | 10.4 | −92.3 (21.2) | 115.4 (26.1) |
| R | −63.2 | 132.4 | −70.7 | 143.4 | −65.4 | 129.1 | −77.4 (31.8) | 158.9 (38.0) | −66.8 | 128.7 | −63.1 | 131.9 | −76.1 (21.2) | 145.9 (26.1) |
| P | −69.0 | 151.2 | −65 | 134.8 | −68.4 (42.4) | 151.2 (49.9) | −67.5 | 153.8 | −68 (31.8) | 152.5 (38.0) | ||||
| A | −69.2 | 153.5 | −65.7 | 153.1 | −69.0 | 149.6 | ||||||||
| P | ||||||||||||||
aX-ray structures.
bPredicted from C13 chemical shifts. Error in brackets.
Fig. 5.
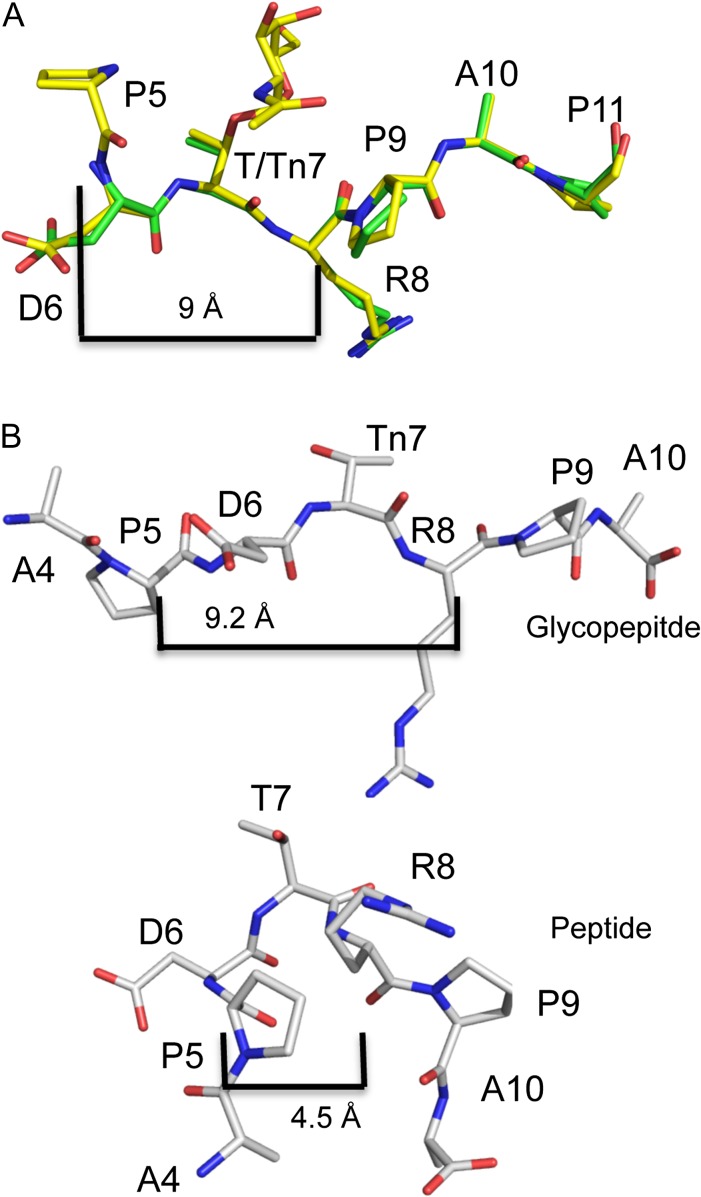
Influence of glycosylation on the conformation of the PDTR region of MUC1. (A) The MUC1 peptide (green) and the glycopeptide (yellow) are in an identical conformation in the combining site of AR20.5. MUC1 is in an extended conformation, with no evidence of a type I β-turn as P5 is 9 Å away from R8. (B) Model of MUC1 glycopeptide and peptide derived from chemical shifts reported in Schuman et al. (2003). Glycosylation of MUC1 with a single GalNAc (Tn7) results in an extended conformation, similar to that observed in the structure of AR20.5. In solution, the unglycosylated peptide adopts a type I β-turn with P5 being 4.5 Å from R8. This figure is available in black and white in print and in color at Glycobiology online.
Both the MUC1 peptide and the glycopeptide are in a nearly identical conformation when seated in the antibody's binding cleft, and show no evidence of a type I β-turn (backbone r.m.s.d. 0.2 Å) (Figures 5A, 6 and Table II). The Pro (P5) that would form the first residue in the turn is disordered in the peptide structure, so the distance to R8 cannot be measured. However, in the glycopeptide, P5 is ordered, and is 9 Å from R8, much greater than 7 Å required for a β-turn. Furthermore, the phi/psi angles of both Asp and Thr do not correspond to those of a type I β-turn (Table II).
Fig. 6.
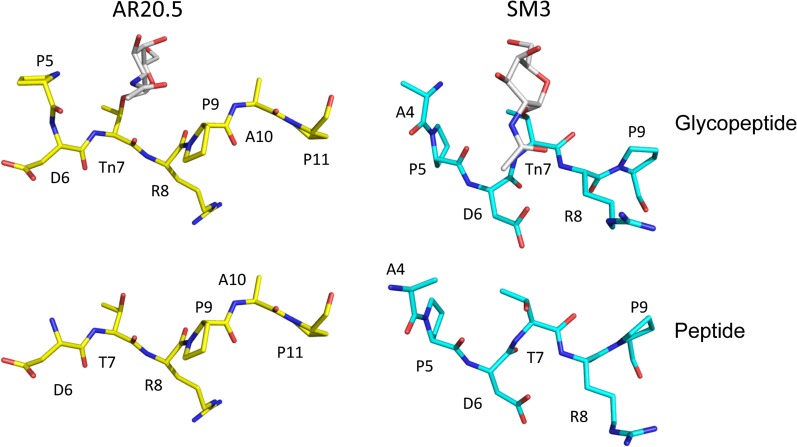
Conformations of MUC1 glycopeptide and peptide bound to AR20.5 (this study) and SM3 (Martinez-Saez et al. 2015). Glycosylation does not influence the conformation of the MUC1 epitope in either antibody (bottom and top panel). The conformation of the antigens is not identical in the two antibodies, but is in a similar extended conformation with no evidence of a turn (left and right panels). This figure is available in black and white in print and in color at Glycobiology online.
A similar trend has been observed in the X-ray structures of SM3 in complex with unglycosylated and glycosylated MUC1 peptides. There is no evidence for the PDTR region adopting a type I β-turn, with the antigen exhibiting an extended conformation (Dokurno et al. 1998; Martinez-Saez et al. 2015) (Table II, Figure 6). Thus, given the available evidence, it appears that in at least in the case of AR20.5 and SM3, the immunodominant conformation of the PDTR region of MUC1 is not a type I β-turn but rather an extended unstructured conformation.
High affinity binding to AR20.5 is mediated by conformational selection: glycosylation of MUC1 alters the conformational equilibrium favoring the extended form of the antigen
An alternative hypothesis to explain the enhanced binding of MUC1 antibodies (including AR20.5) to Tn-glycosylated MUC1 is that the addition of a GalNAc carbohydrate in the DTR motif directly alters the conformational equilibrium of the epitope, shifting the predominant population in solution toward the bioactive conformation recognized by the antibody. If glycosylation of the PDT region of the epitope shifted the conformational equilibrium of the MUC1 VNTR away from a type I β-turn and toward a more extended conformation, then the apparent affinity of AR20.5 for the glycosylated form of the epitope would be increased. There is compelling evidence that this is indeed the case.
The CD spectra of the 8-MER MUC1 peptide and Tn glycopeptide in this study support the hypothesis that glycosylation alters the conformational equilibrium of the peptide. In the absence of glycosylation, the spectra displayed a negative peak at 205 nm characteristic of a type I β-turn (Greenfield 2006), while the Tn glycosylation shifts the peak maxima to 196 nm, which is more characteristic of a disordered structure (Greenfield 2006) (Figure 4). Other studies that examined the structural consequences of MUC1 glycosylation arrived a similar conclusion.
A landmark NMR study examining the effects of the GalNAc glycosylation of the DTR region of a 9-mer MUC1 synthetic peptide/glycopeptide (TSAPDT/TnRPA) found that the unglycosylated peptide adopted a type I β-turn, while glycosylation destabilized the β-turn and shifted the equilibrium toward an extended state even in a short peptide fragment (Table II, Figure 5B) (Schuman et al. 2003). Other recent NMR studies examining the consequences of O-glycosylation in the DTR region of MUC1 resulted in similar conclusions, where the addition of GalNAc on Thr stabilized a extended conformation of the MUC1 peptide (Kinarsky et al. 2003; Rangappa et al. 2016). Using the chemical shifts reported by Schuman et al. (2003), the phi and psi angles for the 9-mer peptide and glycopeptide were predicted, and a model of the peptide/glycopeptide was constructed (Table II, Figure 5B). Interestingly, although not identical, the conformation of the GalNAc (Tn antigen) glycosylated form of MUC1 predicted by Schuman et al. (2003) is very similar to that found in the AR20.5 structures reported here, supporting the hypothesis that glycosylation stabilizes the conformation of the peptide recognized by AR20.5. The predicted conformation of the unglycosylated MUC1 peptide with its type I β-turn would be unable to fit into the AR20.5 combining site: the change in direction caused by type I β-turn configuration of the peptide would likely not allow proper engagement of P9 and P11 in their respective binding pockets on the surface of AR20.5 (Figures 2B and 3B). Therefore, a plausible explanation for the apparent higher affinity of AR20.5 for the GalNAc (Tn antigen) glycosylated form of MUC1 over the naked peptide can then be rationalized by a combination of weak hydrophobic contacts to the carbohydrate combined with the conformational equilibrium of the antigen. At equilibrium, the naked, unglycosylated form of the peptide exists predominantly as a type I β-turn which cannot be bound by the antibody, thus lower affinity is observed as the bioactive, extended form of the antigen is not the major conformation in solution. The addition of GalNAc shifts the conformational equilibrium of the antigen toward the extended state (Schuman et al. 2003) favored by the antibody, resulting in higher affinity binding.
Clinical significance
While the therapeutic potential of MUC1 antibodies has long been under investigation, there have been few successes in bringing these antibodies from the laboratory to the clinic. The failure of a phase II clinical trial of the humanized MUC1 antibody huHMFG-1 (AS1402) (Ibrahim et al. 2011) highlights the importance of selecting an appropriate antibody which not only binds the correct target, but also provides a suitable mechanism of action for clinical efficacy. The target and therapeutic mechanism of AR20.5 is fundamentally different from that of huHMFG-1 and point to alternative therapeutic strategies that may prove to be more successful in the clinic. Unlike AR20.5, huHMFG-1 binds equally well to unglycosylated and Tn-glycosylated MUC1(Karsten et al. 2004). Furthermore, the purported mechanism of anti-tumor action is distinct in the two antibodies. huHMFG-1 functions through antibody-dependent cell-mediated cytotoxicity (ADCC) (Snijdewint et al. 2001) while the anti-tumor activity of AR20.5 is mediated by dendritic cell internalization of antigen–antibody complexes and subsequent T-cell activation (Schultes et al. 2005). The preference of AR20.5 for a glycosylation-dependent conformational epitope appears to be linked to its anti-tumor activity. In vaccination studies, Tn-MUC glycopeptides activated T cells through dendritic cell antigen presentation (Ryan et al. 2009), which is consistent with the observation presented here that AR20.5 preferentially binds a conformational neoantigen stabilized by Tn glycosylation of MUC1. Immunotherapeutic approaches that mobilize T cells to target cancer-specific neoantigens have rapidly emerged as a powerful approach to treating malignancies (Schumacher and Schreiber 2015; Desrichard et al. 2016; Lu and Robbins 2016). In particular, the use of immune checkpoint inhibitors has met with stunning success in the treatment of some melanoma cases (Hodi et al. 2010). A limitation on the successful use of immune checkpoint inhibitors is the requirement for preexisting T-cell populations that can recognize cancer neoantigens (Schumacher and Schreiber 2015). This observation points to a role for antibodies similar to AR20.5 in combination immunotherapies. Since the MUC1 neoantigen recognized by AR20.5 ultimately results in the activation of a tumor-specific T-cell population, combining this treatment with checkpoint inhibitors and chemotherapy, which has also improved tumor response to checkpoint inhibitors (Pfirschke et al. 2016), may result in a highly effective combination immunotherapy treatment option.
Conclusions
The weight of evidence presented here suggests that glycosylation of MUC1 enhances the binding of AR20.5 to its epitope by shifting the conformation equilibrium of the antigen to the extended state which is recognized by the antibody, rather than by increasing the antigen–antibody interactions. Given that the majority of MUC1 antibodies raised against tumor-derived immunogens exhibited a similar high affinity dependence on Tn glycosylation in the DTR region (Karsten et al. 2004), it seems highly probable that this is a general mechanism employed by many MUC1-specific antibodies. Future work will continue to explore the hypothesis that the conformational equilibrium of MUC1 fragments is responsible for high affinity antibody binding and examine the generality of this mechanism in other MUC1-specific antibodies.
Materials and methods
Synthesis of peptides and glycopeptides
Synthetic peptide corresponding to the AR20.5 MUC1 epitope sequence (APDTRPAP) was prepared by Genscript Inc. (Piscataway, NJ).
The Tn-MUC1 compound was synthesized by using microwave-assisted solid phase peptide synthesis (MW-SPPS) on Rink Amide AM LL resin (0.05 mmol). The peptide sequence containing first four amino acid residues (RPAP) was assembled on resin support using the fully automated CEM LIBERTY 908505 peptide synthesizer equipped with a UV detector. The resin was then removed from the synthesizer and Tn moiety was installed manually using Nα-Fmoc-Thr-(AcO3-α-D-GalNAc) (67 mg, 0.1 mmol) in dimethylformamide (DMF) (3 mL) and 2-(7-aza-1H-benzotriazole-1yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) (38 mg, 0.1 mmol), 1-Hydroxy-7-azabenzotriazole (HOAt) (13 mg, 0.1 mmol) and N,N-diisopropylethylamine (DIPEA) (39 µL, 0.2 mmol) as activating reagents. The reaction mixture was microwave irradiated for 10 min and completion of coupling was monitored by Kaiser Test. The resin was then returned to automated peptide synthesizer to extend the peptide until alanine residue (APDTnRPAP). The resin was then removed from synthesizer, washed with dichloromethane (DCM) (10 mL × 3) and further steps were performed manually. N-terminus of the peptide was acetylated using acetic anhydride (10% Ac2O, 5% DIPEA in DMF, 5 mL). The resin containing peptide was then treated with 70% hydrazine in methanol solution for 2.5 h to remove acetyl groups on the sugar moiety. The resin was then washed thoroughly with DMF (5 mL × 3), DCM (5 mL × 3) and MeOH (5 mL × 3), and then dried under vacuum. The resin was swelled in DCM (10 mL) for 30 min and then treated with the cleavage cocktail [Trifluoroacetic acid (TFA) 95%, water 2.5% and triisopropylsilyl 2.5%; 10 mL] for 2.5 h. The resin was filtered and washed with TFA (3 mL). The filtrate was concentrated on a rotary evaporator under vacuum till approximately 1/3 of its original volume. The peptide was precipitated using diethyl ether (0°C; 25 mL) and recovered by centrifugation at 2500 r.p.m. for 20 min. The crude glycopeptide was purified by reversed-phase high performance liquid chromatography (RP HPLC) on an Agilent 1100 series system equipped with an autosampler, UV detector and fraction collector with a Phenomenex Jupiter analytical C-18 column using a linear gradient of 0–100% B (acetonitrile 95%, water 5%, TFA 0.1%) in A (water 95%, acetonitrile 5%, TFA 0.1%) over 50 min and appropriate fractions were lyophilized to afford pure compound. HR MALDI-TOF MS calculated for C45H74N13O17 [M+H] 1068.5326; observed, 1068.5485.
Sequencing of AR20.5 variable region light (Vl) and heavy chains (Vh)
AR20.5 secreting hybridoma was generously provided by Dr. Madi Madiyalakan (OncoQuest Inc., Edmonton, Alberta, Canada) (Dippold et al. 1980). Cells were cultured in RPMI 1640 supplemented with l-glutamine (2 mM), fetal bovine serum (25%) and penicillin–streptomycin at 37°C, 5% CO2. Total RNA was isolated from the hybridoma cells using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA was prepared from 1 µg of total RNA using Superscript III reverse transcriptase (Invitrogen). The Vh and Vl were PCR amplified from cDNA using the primer sets described by Brocks et al. (2001). Successful PCR products were purified and blunt cloned into pJET1.2 and sent for DNA sequencing. Germline sequence analysis was carried out using IMGT V-quest (Brochet et al. 2008).
Production of AR20.5 IgG and purification of Fab fragments
AR20.5 hybridoma cells were cultured in a CELLine CL 1000 bioreactor (Integra Biosciences, Hudon, NH) following the manufacturer's instructions. Supernatant was harvested weekly and diluted 1:2 with buffer [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 0.3 M NaCl]. AR20.5 IgG was purified using protein G (GE Healthcare, Pittsburgh, PA) according to the manufacturer's instructions. Purified protein was dialyzed against buffer (20 mM HEPES pH 7.5, 0.15 M NaCl) and diluted to a concentration of 0.7 mg/mL. For the production and purification of Fab fragments, IgG was mixed with papain (Sigma-Aldrich Corp., St. Louis, MO) using a 1:100 (w/w) ratio and digested for 2.5 h at 37°C [20 mM HEPES pH 7.5, 1 mM dithiothreitol (DTT)], the reaction was quenched with iodoacetamide (10 mM). The protein was dialyzed overnight (20 mM HEPES pH 6.5) and the Fab fragments purified using an Enrich S-cation exchange column (Bio-Rad, Hercules, CA).
Affinity determination using ligand-induced fluorescent changes in MST
Binding affinities of AR20.5 Fab for MUC1 peptide and glycopeptide were determined using a Monolith NT.115 (Nanotemper Technologies, München, Germany). Purified Fab was dialyzed against PBS and labeled with the Dylight® 488 Antibody Labeling Kit (Thermo Fisher Scientific, Fermont, CA) and dialyzed against 50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween-20, pH 7.4. Each replicate contained a 16-step serial dilution (1:2) of unlabeled MUC1 peptide (5.5 μM) or glycopeptide (3.75 μM). The samples were loaded into standard capillaries and heated for 30 s, followed by 5 s cooling at 40% laser power. There was a clear ligand concentration-dependent change in fluorescence. When the samples were mixed with sodium dodecyl sulphate (SDS) (2% SDS, 20 mM DTT) and heated (95°C, 5 min) and rescanned for fluorescence, the concentration-dependent change in fluorescence was abrogated. All experiments were performed with three independent replicates. Affinity, KD, was quantified by analyzing the change in fluorescence as a function of the concentration of the titrated peptide/glycopeptide. The fraction of Fab bound was plotted against the concentration of ligand and the curves were analyzed using GraphPad Prism 6.
Crystallization of AR20.5 Fab in complex with MUC1 peptide and glycopeptides
AR20.5 Fab was concentrated to 13 mg/mL and mixed with either peptide or glycopeptide (1 mM) and screened against the Nextal DWBlock PEGs, PEG II screens (Qiagen Inc., Valencia, CA) using a CrystalGryphon (Art Robbins Inc., Sunnyvale, CA). Optimal crystals of AR20.5 in complex with MUC1 peptide were obtained in 12–15% PEG 20000, 0.1 M Tris-HCl, pH 7.8–8.5. Optimal crystals of MUC1 in complex with glycopeptide were obtained in identical conditions.
Data collection, structure determination and refinement
Crystals were dipped in mother liquor supplemented with 25% glycerol and flash frozen in liquid nitrogen for data collection. X-ray data were collected at the Canadian Light Source (Saskatoon, Canada) on beamline 08ID-1. X-ray data were processed using autoprocess (Grochulski et al. 2012). The structure of AR20.5 Fab was solved by molecular replacement with Phaser (McCoy et al. 2007) using the antibody S25-2 (PDB code: 4HGW) (Brooks et al. 2012) as a search model. Manual model fitting to 2Fo-Fc electron density maps was carried out with Coot (Emsley and Cowtan 2004). Restrained refinement including TLS parameters and NCS restrains was carried out with Phenix (Adams et al. 2010). Final statistics for data collection and the models are provided in Table I.
CD spectroscopy of MUC1 peptide and glycopeptide
8-MER peptide and glycopeptide CD spectra were recorded on a JASCO J-815 Spectro polarimeter (Jasco, Eaton, MD). Spectra were recorded between 290 and 1900 nm at 0.5 nm intervals with a time constant of 2 s at 25°C. Data were collected from three separate scans and averaged. A cylindrical quartz cell of path length 0.1 cm was used for the spectral range with the sample concentration of 0.05 mM. Peptide solutions were made in 0.1 M PBS buffer 7.2. The molar ellipticity values were expressed against wavelength in nm using GraphPad.
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 5T6P and 5T78.
Acknowledgements
We would like to thank Dr. Madi Madiyalakan (Quest Pharmatech Inc.) for his generous gift of the AR20.5 hybridoma. We also would like to thank Dr. Krish Krishnan (CSU Fresno, Department of Chemistry) for assistance with the generation of the MUC1 models from published NMR data. We also acknowledge Dr. Chris Nicodemus for critical reading of the manuscript. This research was supported by start-up funds from CSU Fresno. This research was supported by the National Cancer Institute of the US National Institutes of Health (R01CA88986 to G-JB).
Conflict of interest statement
The corresponding author has received a research grant from the company OncoQuest to humanize the antibody AR20.5. The company did not fund the research presented here.
Abbreviations
ADCC, antibody-dependent cell-mediated cytotoxicity; CD, circular dichroism; HATU,2-(7-aza-1H-benzotriazole-1yl)-1,1,3,3-tetramethyluronium hexafluorophosphate; HOAt, 1-hydroxy-7-azabenzotriazole; MST, microscale thermophoresis; PEM, polymorphic epithelial mucin; RP-HPLC, reversed-phase high performance liquid chromatography; VNTR, variable number of tandem repeats.
References
- Abdel-Aal AB, Lakshminarayanan V, Thompson P, Supekar N, Bradley JM, Wolfert MA, Cohen PA, Gendler SJ, Boons GJ. 2014. Immune and anticancer responses elicited by fully synthetic aberrantly glycosylated MUC1 tripartite vaccines modified by a TLR2 or TLR9 agonist. Chembiochem. 15:1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acres B, Limacher JM. 2005. MUC1 as a target antigen for cancer immunotherapy. Expert Rev Vaccines. 4:493–502. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Struct Biol. 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos V, Stojanovska L, Gargosky S. 2015. MUC1 (CD227): a multi-tasked molecule. Cell Mol Life Sci. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos V, Yuriev E, Ramsland PA, Halton J, Osinski C, Li WJ, Plebanski M, Paulsen H, McKenzie IFC. 2003. A glycopeptide in complex with MHC class I uses the GaINAc residue as an anchor. Proc Natl Acad Sci USA. 100:15029–15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Bueti D, Burford B, Allen D, Julien S, Hollingsworth M, Gammerman A, Fentiman I, Taylor-Papadimitriou J, Burchell JM. 2011. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 13:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc M-P, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized VJ and VDJ sequence analysis. Nucleic Acids Res. 36:W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks B, Garin-Chesa P, Behrle E, Park JE, Rettig WJ, Pfizenmaier K, Moosmayer D. 2001. Species-crossreactive scFv against the tumor stroma marker "fibroblast activation protein" selected by phage display from an immunized FAP-/-knock-out mouse. Mol Med. 7:461. [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Schietinger A, Borisova SN, Kufer P, Okon M, Hirama T, MacKenzie CR, Wang L-X, Schreiber H, Evans SV et al. . 2010. Antibody recognition of a unique tumor-specific glycopeptide antigen. Proc Natl Acad Sci USA. 107:10056–10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Wimmer K, Kosma P, Müller-Loennies S, Brade L, Brade H, Evans SV. 2012. Exploring the cross-reactivity of S25-2: complex with a 5,6-dehydro-Kdo disaccharide. Acta Crystallogr F Struct Biol Crystallogr Commun. 69:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchell J, Gendler S, Taylor-Papadimitriou J, Girling A, Lewis A, Millis R, Lamport D. 1987. Development and characterization of breast cancer reactive monoclonal antibodies directed to the core protein of the human milk mucin. Cancer Res. 47:5476–5482. [PubMed] [Google Scholar]
- Carroll WL, Mendel E, Levy S. 1988. Hybridoma fusion cell lines contain an aberrant kappa transcript. Mol Immunol. 25:991–995. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Rha SY, Stephenson J, Schultes BC, Monroe P, Eckhardt GS, Hammond LA, Whiteside TL, Nicodemus CF, Cermak JM et al. . 2004. Phase I trial of a murine antibody to MUC1 in patients with metastatic cancer: evidence for the activation of humoral and cellular antitumor immunity. Ann Oncol. 15:1825–1833. [DOI] [PubMed] [Google Scholar]
- Desrichard A, Snyder A, Chan TA. 2016. Cancer neoantigens and applications for immunotherapy. Clin Cancer Res. 22:807–812. [DOI] [PubMed] [Google Scholar]
- Dippold WG, Lloyd KO, Li LT, Ikeda H, Oettgen HF, Old LJ. 1980. Cell surface antigens of human malignant melanoma: definition of six antigenic systems with mouse monoclonal antibodies. Proc Natl Acad Sci USA. 77:6114–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokurno P, Bates PA, Band HA, Stewart LMD, Lally JM, Burchell JM, Taylor-Papadimitriou J, Snary D, Sternberg MJE, Freemont PS. 1998. Crystal structure at 1.95 angstrom resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition . J Mol Biol. 284:713–728. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Finn OJ, Jerome KR, Henderson RA, Pecher G, Domenech N, Magarian-Blander J, Barratt-Boyes SM. 1995. MUC-1 epithelial tumor mucin-based immunity and cancer vaccines. Immunol Rev. 145:61–89. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Mariappan SVS, Catasti P, Domenech N, Finn OJ, Gupta G. 1995. Structure of a tumor-associated antigen containing a tandemly repeated immunodominant epitope. J Biomol Struct Dyn. 13:245–260. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Tjandra N, Bu D, Ho C, Montelaro RC, Finn OJ. 1993. Biophysical characterization of one-, two-, and three-tandem repeats of human mucin (Muc-1) protein core. Cancer Res. 53:5386. [PubMed] [Google Scholar]
- Glaffig M, Palitzsch B, Stergiou N, Schull C, Strassburger D, Schmitt E, Frey H, Kunz H. 2015. Enhanced immunogenicity of multivalent MUC1 glycopeptide antitumour vaccines based on hyperbranched polymers. Org Biomol Chem. 13:10150–10154. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ. 2006. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 1:2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochulski P, Fodje M, Labiuk S, Gorin J, Janzen K, Berg R. 2012. Canadian macromolecular crystallography facility: a suite of fully automated beamlines. J Struct Funct Genomics. 13:49–55. [DOI] [PubMed] [Google Scholar]
- Gururaja TL, Ramasubbu N, Venugopalan P, Reddy MS, Ramalingam K, Levine MJ. 1998. Structural features of the human salivary mucin, MUC7. Glycoconj J. 15:457–467. [DOI] [PubMed] [Google Scholar]
- Hilgers J. 1998. ISOBM TD-4 International Workshop on Monoclonal Antibodies Against MUC1 – Foreword. Tumor Biol. 19:V. [PubMed] [Google Scholar]
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC et al. . 2010. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 363:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim NK, Yariz KO, Bondarenko I, Manikhas A, Semiglazov V, Alyasova A, Komisarenko V, Shparyk Y, Murray JL, Jones D et al. . 2011. Randomized phase II trial of letrozole plus anti-MUC1 antibody AS1402 in hormone receptor-positive locally advanced or metastatic breast cancer. Clin Cancer Res. 17:6822–6830. [DOI] [PubMed] [Google Scholar]
- Jeffrey GA. 1997. An Introduction to Hydrogen Bonding. New York, Oxford University Press. [Google Scholar]
- Karsten U, Diotel C, Klich G, Paulsen H, Goletz S, Müller S, Hanisch FG. 1998. Enhanced binding of antibodies to the DTR motif of MUC1 tandem repeat peptide is mediated by site-specific glycosylation. Cancer Res. 58:2541–2549. [PubMed] [Google Scholar]
- Karsten U, Serttas N, Paulsen H, Danielczyk A, Goletz S. 2004. Binding patterns of DTR-specific antibodies reveal a glycosylation-conditioned tumor-specific epitope of the epithelial mucin (MUC1). Glycobiology. 14:681–692. [DOI] [PubMed] [Google Scholar]
- Kimura T, Finn OJ. 2013. MUC1 immunotherapy is here to stay. Expert Opin Biol Ther. 13:35–49. [DOI] [PubMed] [Google Scholar]
- Kinarsky L, Suryanarayanan G, Prakash O, Paulsen H, Clausen H, Hanisch F-G, Hollingsworth MA, Sherman S. 2003. Conformational studies on the MUC1 tandem repeat glycopeptides: implication for the enzymatic O-glycosylation of the mucin protein core. Glycobiology. 13:929–939. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan V, Thompson P, Wolfert MA, Buskas T, Bradley JM, Pathangey LB, Madsen CS, Cohen PA, Gendler SJ, Boons GJ. 2012. Immune recognition of tumor-associated mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated MUC1 tripartite vaccine. Proc Natl Acad Sci USA. 109:261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. 2014. Antibody informatics: IMGT, the International ImMunoGeneTics Information System. Microbiol Spectr. 2:AID-0001-2012. [DOI] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J et al. . 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37:D1006–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sejbal J, Kotovych G, Koganty RR, Reddish MA, Jackson L, Gandhi SS, Mendonca AJ, Longenecker BM. 1995. Structurally defined synthetic cancer vaccines: analysis of structure, glycosylation and recognition of cancer associated mucin, MUC-1 derived peptides. Glycoconj J. 12:607–617. [DOI] [PubMed] [Google Scholar]
- Lu YC, Robbins PF. 2016. Targeting neoantigens for cancer immunotherapy. Int Immunol. 28:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Saez N, Castro-Lopez J, Valero-Gonzalez J, Madariaga D, Companon I, Somovilla VJ, Salvado M, Asensio JL, Jimenez-Barbero J, Avenoza A, et al. . 2015. Deciphering the non-equivalence of serine and threonine O-glycosylation points: implications for molecular recognition of the Tn antigen by an anti-MUC1 antibody. Angew Chem Int Ed Engl 54:9830–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Takada W, Igarashi K, Naruchi K, Miyoshi R, Garcia-Martin F, Amano M, Hinou H, Nishimura S. 2014. A straightforward protocol for the preparation of high performance microarray displaying synthetic MUC1 glycopeptides. Biochim Biophys Acta. 1840:1105–1116. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr. 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller H, Serttas N, Paulsen H, Burchell JM, Taylor-Papadimitriou J. 2002. NMR-based determination of the binding epitope and conformational analysis of MUC-1 glycopeptides and peptides bound to the breast cancer-selective monoclonal antibody SM3. Eur J Biochem. 269:1444–1455. [DOI] [PubMed] [Google Scholar]
- Muller S, Alving K, Peter-Katalinic J, Zachara N, Gooley AA, Hanisch FG. 1999. High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J Biol Chem. 274:18165–18172. [DOI] [PubMed] [Google Scholar]
- Muller S, Goletz S, Packer N, Gooley A, Lawson AM, Hanisch FG. 1997. Localization of O-glycosylation sites on glycopeptide fragments from lactation-associated MUC1. All putative sites within the tandem repeat are glycosylation targets in vivo. J Biol Chem. 272:24780–24793. [DOI] [PubMed] [Google Scholar]
- Nicodemus CF. 2015. Antibody-based immunotherapy of solid cancers: progress and possibilities. Immunotherapy. 7:923–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirschke C, Engblom C, Rickelt S, Cortez-Retamozo V, Garris C, Pucci F, Yamazaki T, Poirier-Colame V, Newton A, Redouane Y et al. . 2016. Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity. 44:343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Schultes BC, Liu D, Kuzma M, Decker W, Madiyalakan R. 2001. Characterization of an anti-MUC1 monoclonal antibody with potential as a cancer vaccine. Hybrid Hybridomics. 20:313–324. [DOI] [PubMed] [Google Scholar]
- Rangappa S, Artigas G, Miyoshi R, Yokoi Y, Hayakawa S, Garcia-Martin F, Hinou H, Nishimura S-I. 2016. Effects of the multiple O-glycosylation states on antibody recognition of the immunodominant motif in MUC1 extracellular tandem repeats. Med Chem Commun. 7:1102–1122. [Google Scholar]
- Richardson JS. 1981. The anatomy and taxonomy of protein structure. Adv Protein Chem. 34:167–339. [DOI] [PubMed] [Google Scholar]
- Ryan SO, Vlad AM, Islam K, Gariepy J, Finn OJ. 2009. Tumor-associated MUC1 glycopeptide epitopes are not subject to self-tolerance and improve responses to MUC1 peptide epitopes in MUC1 transgenic mice. Biol Chem. 390:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ, Morley SD, Jackson DE, Price MR, Tendler SJ. 1992. Structural and computational investigations of the conformation of antigenic peptide fragments of human polymorphic epithelial mucin. Biochem J. 284(Pt 1):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes BC, Eng H, Agopsowicz K, Nicodemus CF. 2005. Anti-MUC1 antibody enhanced helper and cytolytic T cell responses with human dendritic cells presenting MUC1 antigen or MUC1-positive tumor cells. Cancer Res. 65:623–623. [Google Scholar]
- Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science. 348:69–74. [DOI] [PubMed] [Google Scholar]
- Schuman J, Campbell AP, Koganty RR, Longenecker BM. 2003. Probing the conformational and dynamical effects of O-glycosylation within the immunodominant region of a MUC1 peptide tumor antigen. J Pept Res. 61:91–108. [DOI] [PubMed] [Google Scholar]
- Snijdewint FG, von Mensdorff-Pouilly S, Karuntu-Wanamarta AH, Verstraeten AA, Livingston PO, Hilgers J, Kenemans P. 2001. Antibody-dependent cell-mediated cytotoxicity can be induced by MUC1 peptide vaccination of breast cancer patients. Int J Cancer. 93:97–106. [DOI] [PubMed] [Google Scholar]
- Thompson P, Lakshminarayanan V, Supekar NT, Bradley JM, Cohen PA, Wolfert MA, Gendler SJ, Boons GJ. 2015. Linear synthesis and immunological properties of a fully synthetic vaccine candidate containing a sialylated MUC1 glycopeptide. Chem Commun (Camb). 51:10214–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uray K, Mizuno M, Inazu T, Goto K, Hudecz F. 2014. The effect of glycosylation on the antibody recognition of a MUC2 mucin epitope. Biopolymers. 102:390–395. [DOI] [PubMed] [Google Scholar]
- Vlad AM, Muller S, Cudic M, Paulsen H, Otvos L, Hanisch FG, Finn OJ. 2002. Complex carbohydrates are not removed during processing of glycoproteins by dendritic cells: processing of tumor antigen MUC1 glycopeptides for presentation to major histocompatibility complex class II-restricted T cells. J Exp Med. 196:1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner GJ. 2007. Monoclonal antibody mechanisms of action in cancer. Immunol Res. 39:271–278. [DOI] [PubMed] [Google Scholar]