Short abstract
Recently, accumulating evidence provides that dysregulation of microRNAs (miRNAs) is considered to play vital roles in tumor progression. Based on microRNA arrays, we found that microRNA-448 (miR-448) was significantly downregulated in breast cancer tissue specimens. In our study, we were in an effort to clarify the function, the direct target gene, and the molecular mechanisms of miR-448 in breast cancer. By quantitative RT–PCR, we analyzed the expression of miR-448 in 16 patients with BC. Overexpression of miR-448 was established by transfecting miR-448-mimics into MDA-MB-231 and MCF-7 cells, methyl thiazolyl- tetrazolium and colony formation assays were performed to evaluate its effects on cell proliferation. We also performed cell migration and invasion assays in breast cells overexpressing miRNA-448. All the results indicated that overexpression of miR-448 in breast cancer cells markedly suppressed cell proliferation, migration, and invasion. Through the quantitative RT–PCR and Western Blots, we also evaluated epithelial–mesenchymal transition. We found that overexpression of miR-448 also downregulated the expression of vimentin, a well-known mesenchymal marker. Meanwhile, the epithelial marker E-cadherin was unregulated, suggesting that miR-448 inhibited epithelial–mesenchymal transition . Bioinformatics assay coupled with Western Blot and luciferase assays revealed that miR-448 directly binds to the 3′UTR of E-cadherin repressor ZEB1/2, resulting in suppression of epithelial–mesenchymal transition in breast cancer cells.
Impact statement
In our study, we revealed that miR-448 played a vital role in breast cancer development and we also uncovered the mechanisms of it. Following is the short description of the main findings:
miR-448 is downregulated in BC.
miR-448 regulates cell proliferation, migration, and invasion in BC.
miR-448 specifically regulates ZEB1/2 through binding to the 3′UTR in BC cells.
miR-448 inhibits cell migration, invasion, and EMT by targeting to the 3′UTR of ZEB1/2.
Keywords: Cancer, cell, disease, microRNA, molecular, oncogenesis
Introduction
Similarly with most other countries, breast cancer (BC) is reported to be the most common cancer in China.1 In the USA, after skin cancer, BC is the second most frequent cancer among women. In 2017, there will be approximately 252,710 new cases of invasive BC are expected to be diagnosed and about 40,610 women in the U.S. are expected to die (data from http://www.breastcancer.org). Too many risk factors may contribute to developing BC, such as female sex, obesity, reproductive factor, and inherited factor.2–4 BC, associated with a variety of changes in expression of related genes, is a complicated and multifactorial genetic disease.
It has been largely accepted that, in the pathogenesis of human cancers, the deregulation and dysfunction of miRNAs play a significant role.5 Cleaved from long hairpin premiRNA(∼100 nucleotides) precursors by cytoplasmic RNase III Dicer, mature miRNAs are a class of short RNA molecules of ∼22 nucleotides in length.6 Aberrant expression of miRNAs may be linked to tumorigenesis since they play a vitally important role in various cellular processes.7–11 Recent advances have made great progress in unraveling the molecular mechanism of BC. It has been reported that various of miRNA are obviously deregulated in human BC tissues.12 miRNAs are heralded to be one set of the novel biomarkers and therapeutic targets for BC.13,14
Two research groups reported that miR-448 was considered to be an EMT-associated miRNA. Down regulation of miR-448 may amplify the positive feedback loop between NF-κB and miR-448, leading to the induction of EMT and eventually accelerates BC formation.15,16
In the progression toward cancer metastasis, epithelial-to-mesenchymal transition (EMT) is deemed to be a crucial event, which triggers cellular mobility and then results in the invasion of tumor cells.17,18 During the EMT progression, mediated by EMT-inducing transcriptional repressors ZEB1/2(zinc finger E-box binding homeobox), epithelial cells loss of expression of E-cadherin and cell–cell contacts, change their apical-basal polarity, and then transdifferentiate into mesenchymal cells. During the EMT event, the eminent marks are loss of the expression of E-cadherin, the epithelial markers, and increase in the expression of N-cadherin and vimentin which are the mesenchymal markers.19 Recently, it has been reported that by interacting with the miRNA-200 family, the EMT-activator ZEB1/2 plays crucial role in promoting the metastasis.20–22 Thus, targeting the network of ZEB1/2 and miRNAs might shed a new light on the etiology of the disease for fatal tumors, such as BC.
In our study, we find that the expression of miR-448 is downregulated in BC tissues and cell lines, which is correlated with an upregulation of ZEB1/2 in BC cell lines. Furthermore, ectopic expression of miR-448 regulates EMT in BC cells as well as inhibits migration and invasion of BC cells. The effects of miR-448 downregulation on EMT markers and cell mobility are released by deleting the 3′UTR of ZEB1/2. Our study has proved it strongly that miR-448 facilitates the invasive abilities of BC cells by directly interacting with the E-cadherin repressor ZEB1/2.
Materials and methods
Microarrays analysis
Three pairs of breast cancer tissue specimens were subjected to miRNA microarray analysis as previously described.23
Human samples and cell lines
Primary human BC and normal samples were collected at the Affiliated Hospital of Nantong University. All BC cell lines were purchased from the ATCC (Manassas, Virginia, USA). MCF-7 was maintained in RPMI, 1640 supplemented 10% calf serum (CS) and 1% antibiotics (penicillin/streptomycin); MDA-MB-231 was maintained in L-15 supplemented 10% fetal bovine serum (FBS) and 1% antibiotics (penicillin/streptomycin).
Plasmids construction
3′-UTR of ZEB1/2 containing miR-448 response element was inserted into the pGl4.13 luciferase reporter plasmid as described previously.24 The mutated plasmids were generated by PCR using QuickChange XL Site-Directed Mutagenesis Kit (Stratagene).
Cell proliferation assay and colony formation assay
BC cells/well (5 × 103) was seeded in 96-well plates. After overnight incubation, methyl thiazolyltetrazolium (MMT) metabolism test was performed to detect cell proliferation. For the colony formation assay, 500 cells were plated in 60 mm petri dish and cultured for 14 days. Cell staining and colony counting were performed as previously described.25
RNA oligonucleotides and transfection
Control miRNA or miR-448 (50 nM, BiomicsRNAi, Nantong, China) was transfected into BC cells using Lipofectamine RNAi MAX (Invitrogen).
Migration assay and invasion assay
To assess the migration, 5 × 104 BC cells were seeded into the upper chamber of inserts (BD Biosciences), while 1 × 105 BC cells were used for invasion assay. Cell staining and data analysis were performed as previously described.26
Quantitative real-time PCR
Total RNA from BC cells was extracted using TRIzol reagent (Invitrogen) and the cDNA was synthesized by RT-PCR. SYBR green qPCRs were performed on PCR Lightcycler 480 (Roche).
Primer sequences: ZEB1 forward 5′-GATGATGAATGC GAG TCAGATGC-3′, reverse 5′-CTGGTCCTCTTCAGGT GCC-3′; ZEB2 forward 5′-AACAACGAGATTCTACAAG CCTC-3′, reverse 5′-TCGCGTTCCTCCAGTTT TCTT-3′; β-actin forward 5’- CACTCTTCCAGCCTTCCTTC -3′, reverse 5′-GTACAGGTCTTTGCGGATGT -3′. To determine the expression levels of miRNA, hydrolysis Probes miRNA assays (Applied Biosystems) were performed and U6 snRNA was used for normalization.
Western blotting analysis
Cells were lysed in RIPA buffer as previously described26 and the whole cell lysis was subjected to BCA protein assay (Pierce) to evaluate the protein concentration; 20∼50 μg of lysate protein were further separated in SDS-PAGE. All the antibodies used were purchased from Cell Signaling Technology Company. β-actin was used as the endogenous control.
Statistical analysis
For all the experiments, at least three independent experiments were carried out and the results were presented as mean ± SD. Student t-test was employed to assess the differences and statistical significance was defined as p < 0.05.
Results
miR-448 is downregulated in BC tissue specimens and cell lines and has a negative correlation with the level of ZEB1/2
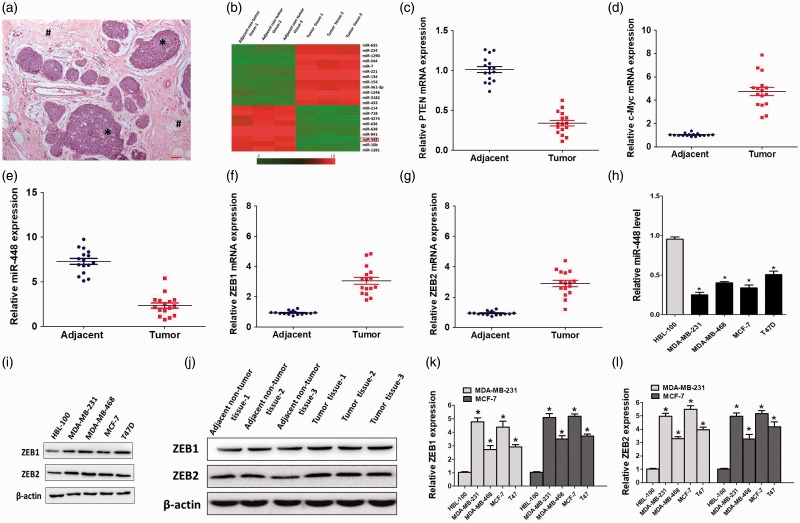
Through miRNA array, we found that comparing with the adjacent non-tumor tissues (<2 cm from tumor margin) (Figure 1(a)), the level of miR-448 was significantly reduced in breast cancer (BC) tissue specimens (Figure 1(b)). The association between miR-448 expression and clinicopathological characteristics of patients is shown in Table 1.5,11 Then, we used paired human BC tissues (tumor tissue and adjacent non-tumor tissues) to demonstrate the level of expression of genes such as tumor suppressors pten and oncogene c-Myc (Figure 1(c) and (d)). The results revealed that the adjacent non-tumor tissues were suitable for standing the normal tissues in our following analysis. It is recognized that miRNAs mainly bind to the 3′ UTR of their target mRNA to inhibit it, and we searched the online database (www.targetscan.org, www.microrna.org.org) for predicting the miRNA target and picked up ZEB1/2 as the candidates (Figure 3(a)).27,28 We then investigated the expression of miR-448 and ZEB1/2 in 16 paired clinical human BC tissues by qRT-PCR. Compared with normal tissues, miR-448 in the tumor tissue was significantly reduced, while the ZEB1/2 increased dramatically (Figure 1(e) to (g)). We further detected the miR-448 level in four human BC cell lines. Compared with the normal human breast cells, BC cells harbored obviously lower miR-448 level (Figure 1(h)). IWB assay results were quantitation and showed that the level of ZEB1/2 was higher in BC cells compared with the normal breast cell (Figure 1(i) to (l)). Hence, miR-448 is downregulated in BC and has a negative correlation with the level of ZEB1/2, indicating an inhibitory effect of miR-448 in BC pathogenesis.
Figure 1.
miR-448 was down-regulated in breast cancer cell lines and tissues.(a) H&E image showed the BC tissues (*) and the adjacent non-tumor breast tissues (#) in patient samples. (c and d) qRT-PCR analysis of pten and c-Myc expression in the human BC tissues and the adjacent non-tumor tissues.(e–g) miR-448 and ZEB1/2 expression in 16 human BC tissues and paired adjacent normal tissues. The average expression was normalized by U6 expression. Each bar represents the mean of three independent experiments. *P < 0.05. (H)qRT-PCR analysis of relative miR-448 expression in normal breast cells and various tumor cell lines. (i– l) Western blotting analysis of ZEB1/2 in normal breast cells, breast cancer cells, adjacent non-tumor tissues and BC tissues, β-actin served as the loading control.
Table 1.
Association between miR-448 expression and clinicopathological characteristics of patients.
| Variables |
Cases (n = 16) |
miR-448 |
P* | ||
|---|---|---|---|---|---|
| n | % | High (5) | Low (11) | ||
| Age (years) | |||||
| ≦ 60 | 9 | 56.25 | 3 | 6 | 0.151 |
| > 60 | 7 | 43.75 | 2 | 5 | |
| ER status | |||||
| Positive | 7 | 43.75 | 3 | 4 | 0.256 |
| Negative | 9 | 56.25 | 3 | 5 | |
| PR status | |||||
| Positive | 9 | 56.25 | 3 | 6 | 0.172 |
| Negative | 7 | 43.75 | 3 | 4 | |
| HER2 status | |||||
| Negative | 7 | 43.75 | 3 | 4 | 0.256 |
| Positive | 9 | 56.25 | 3 | 5 | |
| Lymph node metastasis | |||||
| Yes | 5 | 31.25 | 1 | 4 | 0.032 |
| No | 11 | 68.75 | 3 | 8 | |
| TNM-stage | |||||
| I+II | 10 | 62.50 | 3 | 7 | 0.018 |
| III | 6 | 37.50 | 2 | 4 | |
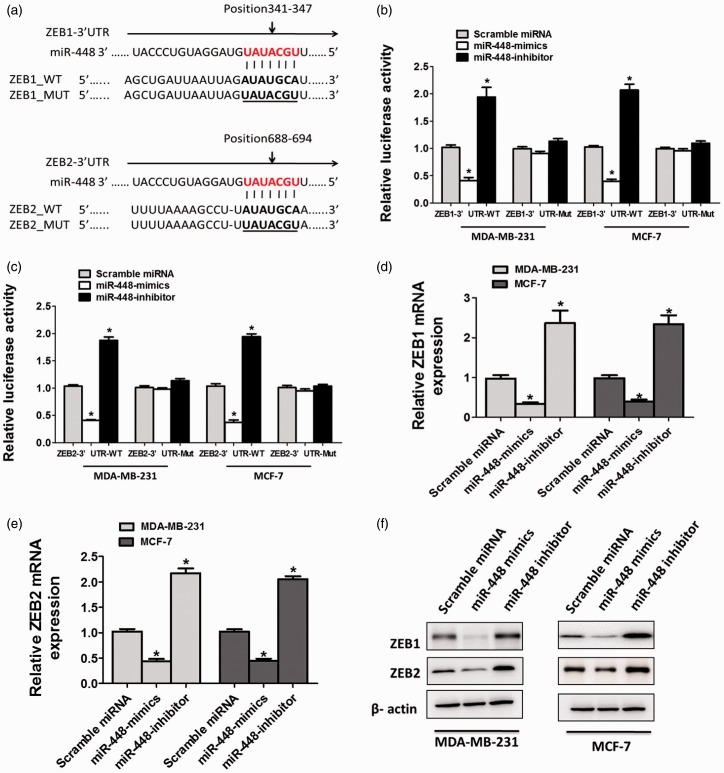
Figure 3.
miR-448 targets ZEB1 and ZEB2 directly in breast cancer cell lines. (a) Putative miR-448 binding sites in the 3′-UTR sequence of ZEB1 and ZEB2. (b) Luciferase activity of MDA-MB-231 or MCF-7 cells transfected with plasmids carrying a wild-type or mutant 3′UTR of ZEB1, in response to miR-448 mimics or inhibitor. (c) Luciferase activity of MDA-MB-231 or MCF-7 cells transfected with plasmids carrying a wild-type or mutant 3′UTR of ZEB2, in response to miR-448 mimics or inhibitor. (d) mRNA levels of ZEB1 and ZEB2 (e) examined by qRT-PCR in MDA-MB-231 or MCF-7 cells transfected with miR-448-mimics, miR-448-inhibitor, or scramble miRNA. *P < 0.05 compared with the scramble miRNA group. (f) ZEB1 and ZEB2 levels were analyzed by Western blotting in MDA-MB-231 or MCF-7 cells transfected with miR-448-mimics, miR-448-inhibitor, or scramble miRNA. (A color version of this figure is available in the online journal.)
miR-448 suppresses the proliferation, migration, and invasion of BC cells
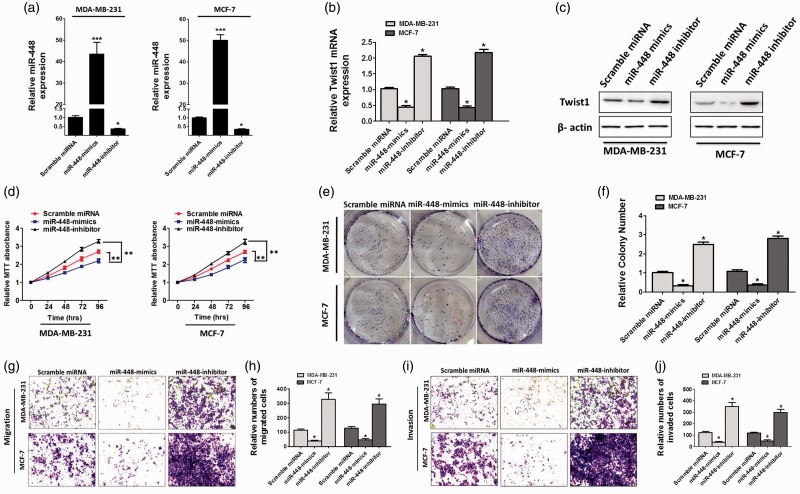
To determine whether decreased miR-448 was related to the occurrence of BC cancers, we generated miR-448-mimics and miR-448-inhibitor. By qRT-PCR, we first confirmed the expression efficiency of miR-448-mimics and miR-448-inhibitor in two independent BC cell lines (MDA-MB-231 and MCF-7) (Figure 2(a)). Next, we examined the level of Twist1, which is also a well-known repressor of E-cadherin and a direct target of miR-448. The results revealed that the Twist1 expression level was downregulated in miR-448-mimics-transfected cells, while upregulated in miR-448-inhibitor-transfected cells (Figure 2(b) and (c)) which means that miR-448-mimics/inhibitor achieves a steady-state efficiency. As evidenced by MTT assay, overexpression of miR-448 effectively inhibited the proliferation of MDA-MB-231 and MCF-7 cells (Figure 2(d)). These results which are confirmed by the colony formation assay (Figure 2(e) and (f)) indicated that the cell proliferation and viability were notably changed after manipulating miR-448 expression levels in BC cells. It has been noted that the EMT is engaged in cell migration and invasion.29,30 Our wound-healing assay results showed that overexpression of miR-448 contributse to a significant reduction of migrating cell numbers in BC cells (Figure 2(g) and (h)). Otherwise, transwell assay revealed that forced expression of miR-448 obviously decreased cell invasion when compared with scramble miRNA-transfected cells (Figure 2(g) and (h)). Conversely, inhibition of miR-448 undoubtedly promoted migration and invasion of BC cells (Figure 2(g) to (j)).
Figure 2.
Ectopic expression of miR-448 suppresses proliferation, colony formation, migration, and invasion in breast cancer cells in vitro. (a) miR-448 levels in MDA-MB-231 and MCF-7 cells transfected with scramble miRNA, miR-448-mimics, or miR-448-inhibitor. (b and c) qRT-PCR analysis of relative TWIST1 expression in these breast cancer cells after treating with miR-448 mimics or inhibitors. (d) MTT assay of MDA-MB-231 and MCF-7 cells transfected with scramble miRNA, miR-448-mimics, or miR-448-inhibitor. (e and f) Representative images and quantification of the colony formation assay. (g and i) Migration and invasion assay of MDA-MB-231 and MCF-7 cells transfected with scramble miRNA, miR-448-mimics, or miR-448-inhibitor. (h and j) The cell number of migrated cells were counted in randomly selected fields and presented in bar graph (means ± SD; *P < 0.05, Student’s t test). (A color version of this figure is available in the online journal.)
miR-448 regulates ZEB1/2 through direct binding to the 3′UTR in BC cells
miRanda and TargetScan analysis predicted one same binding site in the 3′UTR of ZEB1/2, suggesting that miR-448 may directly target ZEB1/2 (Figure 3(a)). To confirm the interaction between miR-448 and ZEB1/2, we constructed luciferase reporter plasmids which contain wild-type 3′-UTR ofZEB1/2 or miR-448 response element mutant (MUT) sequences (Figure 3(a)). Co-transfection of ZEB1–3′UTR-WT and miR-448-mimics into BC cells resulted in dramatically lower luciferase activity than co-transfection with scramble miRNA and this reduction would be rescued in ZEB1–3′UTR-MUT or miR-448-inhibitor-transfected cells, suggesting that miR-448 directly targets ZEB1in (Figure 3(b)). Likewise, co-transfection of ZEB2–3′UTR-WT and miR-448 resulted in much lower luciferase activity than co-transfection with scramble miRNA and recovered to the equal activity in ZEB2–3′UTR-MUT or miR-448-inhibitor-transfected cells, suggesting that miR-448 directly targets ZEB2 (Figure 3(c)). Same results were obtained in BC cell lines (Figure 3(b) and (c)). To test whether miR-448 is an endogenous regulator of ZEB1/2, 48 h after miR-448-mimics or miR-448-inhibitor transfection, BC cells were collected to analyze the mRNA and protein level of ZEB1/2. The results showed that ZEB1/2 mRNA and protein level in BC cells were markedly downregulated after overexpression of miR-448 and the inhibitory effects would be ablated when the expression of miR-448 was inhibited (Figure 3(d) to (f)). Collectively, miR-448 in deed downregulates ZEB1/2 through directly targeting the 3′UTR region.
miR-448 inhibits EMT, cell migration, and invasion by targeting complementary sites in the 3-UTR of ZEB1/2
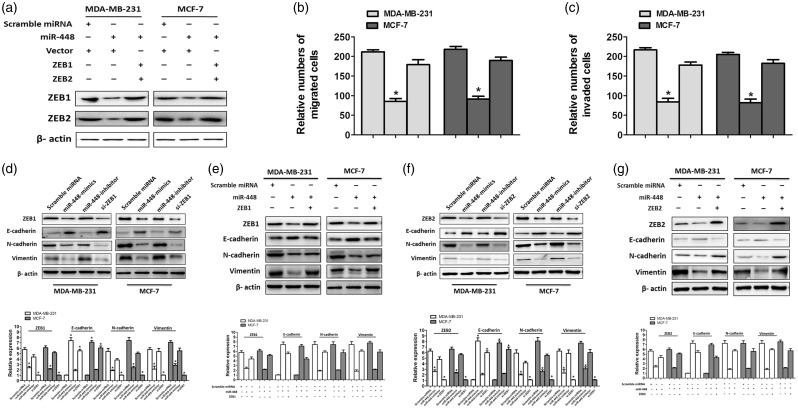
ZEB1 and ZEB2, which are well recognized as important regulators of EMT in BC,21,22 were confirmed to be the targets of miR-448(Figure 3). We transfected scramble miRNA and miR-448-mimic into two BC cell lines separately and subjected transfected cells to WB to detect ZEB1/2. WB results indicated that ZEB1/2 level was reduced by overexpression of miR-448 (Figure 4(a)). In addition, migration and invasion ability of BC cells were evidently reduced when we overexpressed the miR-448, while this inhibitory effects would be released when we co-transfected with ZEB1 or ZEB2 (Figure 4(b) and (c)). Resembling the inhibitory effects of si-ZEB1 or si-ZEB2, overexpression of miR-448 upregulated E-cadherin levels, while N-cadherin and vimentin, the mesenchymal markers, were downregulated (Figure 4(d) and (f)). In contrast, cells co-transfected with miR-448-mimics and 3′ UTR deleted ZEB1 appeared to have a nearly identical level of EMT markers to the scramble miRNA-transfected control cells (Figure 4(e)). Similar results obtained from the cells co-transfected with miR-448-mimics and 3′ UTR deleted ZEB2 (Figure 4(g)). Therefore, we identified that, through targeting to the 3′UTR of ZEB1/2, miR-448 functions as a tumor suppressor in BC cells.
Figure 4.
miR-448 regulating cell migration, invasion, and epithelial-mesenchymal transition related molecules through targeting the 3′UTR ofZEB1/2 in breast cancer cells. (a) Western blotting analysis of ZEB1 and ZEB2 expression in MDA-MB-231 and MCF-7 cells transfected with indicated molecules. β-actin was used as a loading control. (b) Migration assay to determine the migration abilities of MDA-MB-231 and MCF-7 cells transfected with indicated molecules. (c) Cell invasion assay to assess the invasion abilities of MDA-MB-231 and MCF-7 cells transfected with indicated molecules.*P < 0.05 compared with scramble miRNA and vector transfected cells. (d and f) MDA-MB-231 and MCF-7 cells were transfected with miR-448 mimics or inhibitor. si-ZEB1 or si-ZEB2 were used for knockdown of miR-448 target genes, respectively. The protein expression levels of EMT-related molecules were analyzed by immunoblotting. (e and g) Co-transfection of miR-448 with 3′UTR-deleted ZEB1 or ZEB2 plasmid rescued the expressions of EMT-related molecules. The expressions were analyzed by immunoblotting.
Discussion
Accumulating evidence has demonstrated that miRNAs regulate the expression of target genes that involved in BC cells,13,14 which inspire us a new insight to recover the molecular mechanism of BC.11,12 miR-448 has been demonstrated to participate in BC progression.15,16 We employed qRT-PCR for miR-448 expression on 16 paired of BC and matched tumor-adjacent tissues. In this work, we showed that the expression of miR-448 in tumor-adjacent tissues was notably higher than that in BC. In addition, miR-448 was expressed at significantly lower levels in BC cell lines. Besides, our data revealed that deduced expression of miR-448 was in linkage to higher level of ZEB1/2. On the other hand, overexpression of miR-448 restrained proliferation, migration, and invasion of BC cells. On the contrary, knockdown of miR-448 led to the increase of viability and motility of BC cells. Taken together, the results we presented indicated that miR-448 indeed functions as a tumor suppressor in breast cancer.
Our data showed a negative correlation between the expression of miR-448 and the protein level of ZEB1/2 which are the repressors of E-cadherin. It is widely accepted that EMT plays a crucial role in tumor metastasis.17,18 Accordingly, we determined the effect of miR-448 alteration on EMT (marked by E-cadherin and vimentin). The obviously increased expression of E-cadherin and simultaneous decreased protein level of vimentin indicated that miR-448 is a negative regulator of EMT in BC cells. On the contrary, downregulation of miR-448 increased migrated and invaded BC cells. Altogether, miR-448 inhibited migration and invasion of BC cells by suppressing EMT. In addition, through repression of a number of master regulators of epithelial polarity, ZEB1/2 are critical regulators in the expression of E-cadherin and EMT progression.21,22,31–36 By using luciferase reporter assays, we confirmed that miR-448 functioned by directly binding to the 3′UTR of ZEB1/2 transcripts. Furthermore, overexpression of ZEB1/2 results in the ectopic expression of miR-448, and then rescued the migration activity of BC cell lines. Consequently, we propose that disruption of miR-448 may be a critical event in BC metastasis.
In conclusion, we found that downregulation of miR-448 is observed in BC tissues and cells in vitro. The decreased expression of mir-448 is of physiological significance since miR-448 is able to delay the proliferation, migration, and invasion of BC cells by suppressing ZEB1/2-induced EMT. In summary, we consider miR-448 as a tumor suppressive miRNA that function as inhibiting the epithelial-mesenchymal transition in BC cells by directly targeting the E-cadherin repressor ZEB1/2. Thus, for anti-cancer therapy of BC, we believe that miR-448 may be a suitable diagnostic marker and a potential target.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; Peng Ma, Kan Ni, Jing Ke, Wenyi Zhang, Ying Feng, Qinsheng Mao conducted the experiments, Qinsheng Mao wrote the manuscript, and these authors contributed equally to this work: Peng Ma, Kan Ni.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was financially supported by the Nantong Science and Technology Projects (No.MS22015092).
Reference
- 1.Fan L, Strasser-Weippl K, Li J-J, St Louis J, Finkelstein DM, Yu K-D, Chen W-Q, Shao Z-M, Goss PE. Breast cancer in China. Lancet Oncol 2014; 15:e279–89 [DOI] [PubMed] [Google Scholar]
- 2.Protani M, Coory M, Martin JH. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res Treat 2010; 123:627–35 [DOI] [PubMed] [Google Scholar]
- 3.MacMahon B, Cole P, Lin T, Lowe C, Mirra A, Ravnihar B, Salber E, Valaoras V, Yuasa S. Age at first birth and breast cancer risk. B World Health Organ 1970; 43:209. [PMC free article] [PubMed] [Google Scholar]
- 4.Brinton LA, Schairer C, Hoover RN, Fraumeni JF. Menstrual factors and risk of breast cancer. Cancer Invest 1988; 6:245–54 [DOI] [PubMed] [Google Scholar]
- 5.Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012; 482:347–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97 [DOI] [PubMed] [Google Scholar]
- 7.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005; 33:1290–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol 2011; 39:311–8 [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer. The PRIME study. J Clin Oncol 2010; 28:4697–705 [DOI] [PubMed] [Google Scholar]
- 10.Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, Liu X. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer 2013; 13:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834–8 [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065–70 [DOI] [PubMed] [Google Scholar]
- 13.Heneghan H, Miller N, Lowery A, Sweeney K, Kerin M. MicroRNAs as novel biomarkers for breast cancer. J Oncol 2009; 2009:950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015; 5:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Chen Z, Cao X, Xu J, Xu J, Chen Y, Wang W, Chen Q, Tang F, Liu X. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial–mesenchymal transition of breast cancer cells. Cell Death Differ 2011; 18:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mak KK, Wu AT, Lee WH, Chang TC, Chiou JF, Wang LS, Wu CH, Huang CYF, Shieh YS, Chao TY. Pterostilbene, a bioactive component of blueberries, suppresses the generation of breast cancer stem cells within tumor microenvironment and metastasis via modulating NF‐κB/microRNA 448 circuit. Mol Nutr Food Res 2013; 57:1123–34 [DOI] [PubMed] [Google Scholar]
- 17.Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2:442–54 [DOI] [PubMed] [Google Scholar]
- 18.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006; 127:679–95 [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Lamouille S, Derynck R. TGF-β-induced epithelial to mesenchymal transition. Cell Res 2009; 19:156–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, Zur Hausen A. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11:1487–95 [DOI] [PubMed] [Google Scholar]
- 21.Park S-M, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22:894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283:14910–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FT RZ, He Y, Zou M, LG TX. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PloS One 2012; 7:e35435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renthal NE, Chen C-C, Koriand’r CW, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A 2010; 107:20828–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Liu Z, Xu H, Yang Q. miR-409-3p suppresses breast cancer cell growth and invasion by targeting Akt1. Biochem Biophys Res Commun 2016; 469:189. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, PreisM Dragnev KH, Li H, Renzo JD, Bak M, Freemantle SJ, Kauppinen S, Dmitrovsky E. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 2010; 120:1298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010; 11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15–20 [DOI] [PubMed] [Google Scholar]
- 29.Ford CE, Jary E, Ma SSQ, Nixdorf S, Heinzelmann-Schwarz VA, Ward RL. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS One 2013; 8:e54362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Yang M, Gan L, He T, Xiao X, Stewart MD, Liu X, Yang L, Zhang T, Zhao Y. DLX4 upregulates TWIST and enhances tumor migration, invasion and metastasis. Int J Biol Sci 2012; 8:1178–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, Van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 2001; 7:1267–78 [DOI] [PubMed] [Google Scholar]
- 32.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005; 24:2375–85 [DOI] [PubMed] [Google Scholar]
- 33.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007; 26:6979–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szydlowska K, Zawadzka M, Kaminska B. Neuroprotectant FK506 inhibits glutamate‐induced apoptosis of astrocytes in vitro and in vivo. J Neurochem 2006; 99:965–75 [DOI] [PubMed] [Google Scholar]
- 35.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611–22 [DOI] [PubMed] [Google Scholar]
- 36.Ellert-Miklaszewska A, Kaminska B, Konarska L. Cannabinoids down-regulate PI3K/Akt and Erk signalling pathways and activate proapoptotic function of Bad protein. Cell Signal 2005; 17:25–37 [DOI] [PubMed] [Google Scholar]