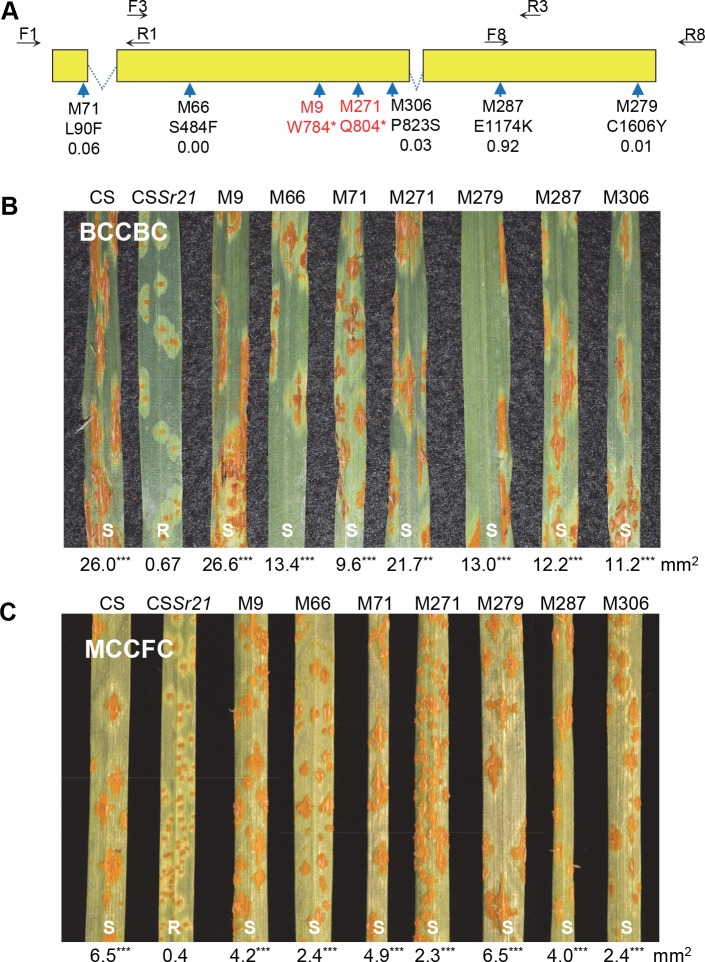
Fig 2. Validation of the Sr21 candidate gene using EMS mutants.
(A) Structure of gene CNL1 (from start codon to stop codon). Primers used for haplotyping are indicated by black arrows. Dotted lines indicate introns, and yellow rectangles indicate coding exons. The positions of the mutations are indicated by blue arrows. Mutation names in red indicate premature stop codons and in black indicate amino acid changes, which are indicated below the names with the corresponding SIFT scores “Sorting intolerant from tolerant” scores lower than 0.05 are predictive of deleterious missense mutations [50]). (B) Infection types on Chinese Spring (CS), CSSr21, and seven mutants inoculated with Pgt race BCCBC. Numbers below leaves indicate average pustule sizes (n = 3) and superscripts indicate significance of differences with CSSr21 (*** = P< 0.001). (C) Same as B above but with race MCCFC.