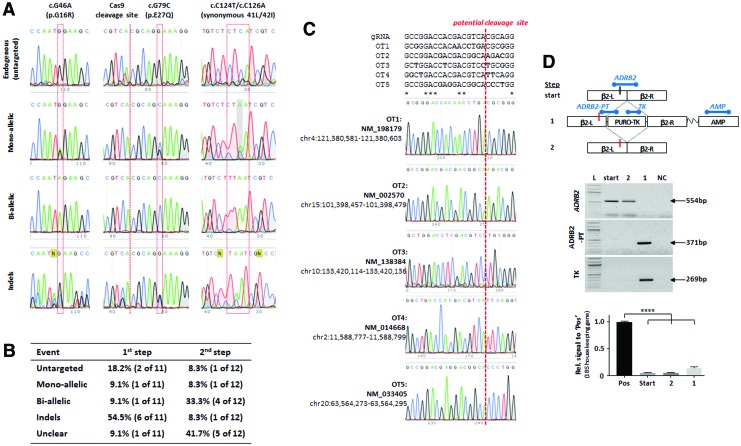
FIG. 3.
Gene editing at the ADRB2 locus in hPSCs. (A) Shows representative chromatogram synopses flanking positions 46, 79, and 124–126 of untargeted, mono- and biallelic targeting, and indels. A complete set for step 1 and step 2 targeting is in Supplementary Figure S2A and B. The table in (B) summarizes the different targeting events identified after step 1 (midpoint; after puromycin selection for clones containing the positive–negative selection cassette) and step 2 (after ganciclovir selection for clones in which the cassette has been excised). In (C), high-risk OT sites were classified as known coding or regulatory sequences where gRNAs had full PAM site complementarily and/or fewer than five mismatches with the target. PCR genotyping showed no evidence for off target events. In (D), random integration was tested. The schematic shows the stages of targeting and location of PCRed regions. ADRB2 is a control for genomic DNA, while ADBR2-PT and TK test for the presence of the targeting cassette; results are shown in the gel images. Since no product was identified for AMP within the pBluescript backbone, qPCR was used and compared against a positive control (pos) comprising plasmid DNA diluted to the equivalent of single-copy gene level in HUES7 parental DNA. Housekeeping gene was 18S, n = 3 ± SD; ****P < 0.001, Dunnett's test. OT, off target; qPCR, quantitative polymerase chain reaction. Color images available online at www.liebertpub.com/scd