Figure 4.
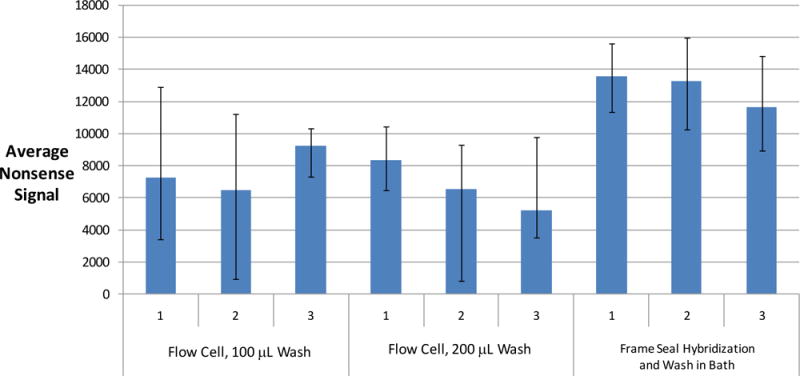
Comparison of dN20 nonsense signals after different washing conditions using the Wound v2.0 array. Six flow cells with a pipettor using two different volumes of 1X SSPE and 25 μL of water versus the gold standard of hybridizing in Frame-Seals and washing arrays in a 1X SSPE microarray bath followed by a water rinse. The flow cells undergo an on-chip amplification procedure whereas the Frame Seals are used only for hybridization using amplified product from a standard PCR tube. The Frame Seals are subsequently removed in order to directly expose the array to the wash buffers in the bath, a water rinse, and air drying. The flow cells are dried per the protocol described in the experimental section and imaged with an Aurora Port Array 5000 for 0.2 seconds. The input sample is 104 genomic copies of S. pyogenes for both the flow cells and the Frame Seals. The nonsense spots are identified with Spotfinder, and the integral fluorescent intensity of each of 4 nonsense spots minus the local background is averaged and plotted along with error bars, which represent the minimum and maximum values. The nonsense signals are inversely proportional to the effectiveness of washing because the dN20 nonsense probes fluoresce due to non-specific binding with labeled product.
