Abstract
MTH1 has become a new rising star in the field of ‘cancer phenotypic lethality’ and can be targeted in many kinds of tumors. This study aimed to explore the anticancer effect of MTH1-targeted drug (S)-crizotinib on osteosarcoma (OS) cells. We detected MTH1 expression in OS tissues and cells using immunohistochemistry and western blot. The effects of MTH1 on OS cell viability were explored using the siRNA technique and CCK8. The anticancer effects of the MTH1-targeted drug (S)-crizotinib on OS cells were explored by in-vitro assays. The intracellular 8-oxo-dGTP level and oxygen reactive species (ROS) of OS cells were detected by Cy3-conjugated avidin staining and dichlorofluorescein diacetate staining, respectively. The expression of MTH1 was significantly higher in OS tissues and cell lines than that in the corresponding adjacent tissues and osteoblastic cell line. The proliferation of OS cells was significantly inhibited through knockdown of MTH1 by siRNA technology. (S)-Crizotinib could inhibit the proliferation of OS cells with an increase in the apoptosis levels and causing G0/G1 arrest by targeting MTH1 and activating ROS. In addition, (S)-crizotinib could inhibit the migration of OS cells. (S)-Crizotinib could suppress the proliferation and migration, cause G0/G1 arrest, and increase the apoptosis level of OS cells by targeting MTH1 and activating ROS. This study will provide a promising therapeutic target and the theoretical basis for the clinical application of (S)-crizotinib in OS.
Keywords: (S)-crizotinib, MTH1, osteosarcoma, 8-oxo-dGTP, targeted therapy
Introduction
Osteosarcoma (OS) is the most common primary bone malignancy. Disease-free survival was increased from less than 20% before the introduction of effective chemotherapy to 55–75% and overall survival to 85%. However, the survival rate of OS patients has almost not altered in the past 30 years because of no remarkable improvement in therapeutic strategies 1. Thus, new therapeutic strategies are urgently needed to improve the prognosis of OS patients. Targeted therapy has achieved great success in various tumors. These treatments are dominated by targeting the genetic defects found in cancers, such as oncogenes (e.g. imatinib for BCR-ABL mutated cancer) or nononcogenic genetic defects (e.g. PARP inhibitors for BRCA1 and BRCA2 mutated cancer 2). However, it is disappointing that targeting genetic defects in a personalized strategy is limited by the high degree of intratumor heterogeneity, adaptation of genetic networks, and high somatic mutation rates in many cancers 3.
Tumor cells suffer constant attacks on DNA by endogenous and environmental stimuli; thus, genomic instability is an underlying hallmark of cancers. To deal with the adverse effects of DNA damage, cells have developed many mechanisms, collectively termed the DNA-damage response (DDR), to detect DNA lesions, signal their presence, and promote their repair 4. Loss of one or more DDR pathways, increased replication stress, and higher levels of endogenous DNA damage are all differentiating aspects of cancer DDR that can be targeted therapeutically. Drugs targeting DDR pathways are predicted to preferentially kill cancer cells and spare normal cells, providing significant benefits for patient over conventional cancer chemotherapeutic approaches 5.
A high level of reactive oxygen species (ROS) is a general phenotype and has been detected in almost all cancers, where they play an important role in regulating tumor proliferation, development, and metastasis 6–8. ROS can cause oxidative damage in double-stranded DNA directly, or to free bases in the cellular and mitochondrial deoxynucleoside triphosphate pool, whereas the free deoxynucleoside triphosphate precursor pool is 190–13 000 times more susceptible to damage 9. A major product of ROS damaging DNA is 8-oxo-7,8-dihydroguanine triphosphates (8-oxo-dGTP) 10,11. The incorporation of 8-oxo-dGTP into DNA can cause G : C–T : A mispairing mutations, which are considered to have a close relationship with the development and progression of tumors, cell aging, and some degenerative diseases 12. MTH1 (MutT Homolog 1) is a kind of hydrolase that selectively degrades 8-oxo-dGTP, thus avoiding mispairing mutations and cell death 9,13,14. MTH1 has been reported to be highly expressed in many kinds of tumors and play a vital role in the survival of tumor cells 15–21. MTH1-targeted inhibitors can suppress the proliferation of tumor cells and induce cell apoptosis; thus, MTH1 inhibitors are a novel anticancer strategy 22–24. Moreover, targeting MTH1, a common cancer phenotype, may overcome the problem of intratumor heterogeneity and may be applied to a wider range of tumors 22.
For the special structure and internal environment homeostasis in bone tumors, an increase in ROS level has been observed in many researches 25,26. Recent studies have shown that knockdown of MTH1 gene expression in U2OS cell lines can cause marked damage in DNA and inhibit cell proliferation 22,27. Some other studies showed that inhibition of MTH1 induces apoptotic cancer cell death in MG63 28. The above researches provided evidence that the repair of DNA oxidative damage plays a major role in OS cell survival and MTH1 may become an underlying therapeutic target for OS. Yet, to the best of our best knowledge, the expression and effect of MTH1 on OS have never been reported. Therefore, we use immunohistochemistry (IHC) to detect the expression of MTH1 in OS tissues and analyze its correlation with clinical prognosis. To further explore the application value of MTH1 as a therapeutic target for OS, we decreased the gene expression of MTH1 and inhibited MTH1 with targeted drugs to investigate the effects on OS cell lines. Several MTH1 inhibitors have been explored in preclinical studies 23,24,27,28. Among these inhibitors, (S)-crizotinib, the (S)-enantiomer of crizotinib [a targeted drug applied to the treatment of EML4-ALK-positive non-small-cell lung cancer (NSCLC)], has greater potential for use in clinic because of its pharmacokinetic and pharmacodynamic properties 23. Recently, (S)-crizotinib was also shown to induce ROS in human NSCLC cells 29, which would induce oxidative damage and cell apoptosis. All these taken together would enhance the anticancer effect of (S)-rizotinib. Therefore, in this study, we used (S)-crizotinib to investigate its anticancer effects on OS cell lines by in-vitro assays. The results of our study showed that (S)-crizotinib could inhibit the proliferation of OS cells by increasing the apoptosis levels and causing G0/G1 arrest by targeting MTH1 and activating ROS. In addition, (S)-crizotinib could inhibit the migration of OS cells. This study could provide a promising therapeutic target; and the theoretical basis for the clinical application of (S)-crizotinib in OS.
Patients and methods
Patients and tissues
Thirty-one patients with OS were selected from the files of the Department of Pathology of Wuhan Union Hospital, Tongji Medical University, Huazhong University of Science and Technology, China. All patients underwent operations between 2012 and 2015 in the Orthopedic Hospital of Union Hospital, and from their paraffin-embedded surgical samples, we obtained 31 OS tissue samples and 16 corresponding adjacent normal tissue samples (para-carcinoma tissues; 15 paraffin-embedded samples had no adjacent normal tissues). The diagnoses of each patient were confirmed by the OS pathologist and the pathological types included 24 conventional OS, 11 chondroblastic OS, two parosteal Oss, and three telangiectatic OS. The patients ranged in age from 11 to 68 years (median: 20 years). Twenty-one males and 10 females were included in the study population. The correlations of clinicopathological parameters (including age, sex, and histological subtype) with MTH1 expressions in OS are shown in Table 1. All cases were reviewed independently by two pathologists and discrepancies were resolved by consensus review. Our study protocol was reviewed and approved by the Wuhan Union Institutional Review Board and informed consent was obtained from each patient.
Table 1.
Correlation between the expression of MTH1 and clinicopathological features in osteosarcoma
Immunohistochemistry
Immunohistochemical stainings were carried out using 4-μm tissue sections according to standard procedures. Briefly, after antigen retrieval, samples were blocked with 3% peroxidase-blocking solution (10011218; Sinopharm, Beijing, China) for 20 min at room temperature and primary antibodies against MTH1 (ab200832; Abcam, Shanghai, China) were then incubated with samples overnight (antibody against MTH1 was used at 1 : 50 dilutions). Slides were washed with Tris-buffered saline (TBS) and the corresponding secondary antibody was incubated with the sample for 30 min at room temperature. Samples were developed using 3,3′-diaminobenzidine (K5007; Dako, Glostrup, Denmark), counterstained with hematoxylin, and mounted according to the instructions provided by the manufacturer.
The staining intensity of the cells was graded as follows: 0, negative; 1, light yellow; and 2, deep yellow or brown. Grading according to the ratio of positive cells was performed as follows: 0, ratio of positive cells≤5%; 1, 5%<ratio of positive cells≤25%; 2, 25%<ratio of positive cells<50%; and 3, ratio of positive cells≥50%. The final score for the expression of MTH1 was determined through the multiplication of the score for staining intensity by the score for the ratio of positive cells. The expression levels were graded as follows: −, scores≤1; +, 2≤scores≤3; ++, 4≤scores≤5; and +++, scores≥6. The expression levels – and + were determined as MTH1 negative, whereas ++ and +++ were determined as positive.
Cell culture
Human OS cell lines U2OS, MG63, Saos-2, and MNNG/HOS and osteoblastic cell line hFOB1.19 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultivated in suitable essential medium [MG63 and MNNG/HOS cell lines in Eagle’s minimum essential medium (Gibco, Grand Island, New York, USA); hFOB1.19 in Dulbecco’s modified Eagle’s medium/ham’s F-12 (Gibco) containing 0.3 mg/ml G418; U2OS in RPMI medium 1640 (Gibco); and Saos-2 in McCOY’s 5A medium (Gibco)] supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 units/ml penicillin, and 100 mg/ml streptomycin (Gibco). All cells were incubated at 37°C and 5% CO2 in an incubator.
Western blot
Cells of various groups were washed with ice-cold PBS. RIPA lysis buffer [50 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS] (Beyotime Biotechnology, Haimen, People’s Republic of China) plus phenylmethanesulfonyl fluoride (Beyotime Biotechnology) were used for total protein extraction from OS cell lines. The Enhanced BCA Protein Assay Kit (Beyotime Biotechnology) was used to detect the protein concentration. Equivalent amounts of protein (40 µg) were loaded on 10 or 15% SDS-PAGE gels and then transferred to polyvinylidene fluoride membranes (BioRad, Hercules, California, USA). The membranes were blocked with 1% nonfat milk in a mixture of TBS and Tween-20 for 1 h at room temperature and incubated overnight at 4°C with the primary antibodies against MTH1 (ab200832; Abcam) and β-actin (Sungene Biotech, Tianjin, China). After incubation with secondary antibodies (Santa Cruz Biotechnology, Dallas, Texas, USA) labeled with horseradish peroxidase for 1 h at room temperature, immunoreactive bands were developed using BeyoECL Plus (Beyotime Biotechnology).
siRNA experiments
MTH1 siRNA and nontargeting (NT) siRNA were designed and synthesized by GenePharm (Shanghai, China). NT RNA was used as a control and the MTH1 siRNA sequence was GACGACAGCUACUGGUUUC. One day before transfection, cells were seeded in medium without antibiotics such that they would be 30–50% confluent at the time of transfection. Transfections were performed with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. The medium was changed with complete growth medium 24 h after transfection.
Real-time PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen). Gene expression was detected by real-time PCR with All-in-One qPCR Master Mix (GeneCopoeia, Rockville, Maryland, USA) using the ABI stepone plus Sequence Detection System (Applied Biosystems, Foster City, California, USA). The following primer sequences were used for real-time PCR: 5′-GTGCAGAACCCAGGGACCAT-3′ (forward) and 5′-GCCCACGAACTCAAACACGA-3′ (reverse) for MTH1. Results were quantified using the  method with β-actin expression levels for normalization.
method with β-actin expression levels for normalization.
Cell counting kit-8 assay
For the study of transfection, cells were seeded in 96-well cell culture plates and transfected with MTH1 siRNA or NT siRNA as described above. For the study of (S)-crizotinib (Apexbio, Houston, Texas, USA), cells were seeded in 96-well plates with a density of 3–5×103 cells/well, 24 h after which the original culture medium was changed with the culture medium plus (S)-crizotinib alone at different concentrations or 5 µmol/l (S)-crizotinib combined 5 mmol/l anti-oxidant N-acetyl-l-cysteine (NAC; Beyotime Biotechnology) or 0.5 µl of dimethyl sulfoxide (DMSO) for the control group. After 1–5 days, culture medium was changed with 10 µl of CCK8 solution (Dojindo, Kumamoto, Japan) in each well and the plates were incubated for 3 h at 37°C according to the manufacturer’s instructions. The absorbance of each well was determined using a microplate reader (Bio-Tek, Winooski, Vermont, USA) at 450 nm.
Colony formation assay
The cell survival of OS cells was measured by the colony formation assay. Cells were trypsinized into a single-cell suspension and were seeded in six-well plates at a density of 1×103 cells/well. The medium was changed with the 2 ml culture medium plus (S)-crizotinib at different concentrations or 10 µl of DMSO as a control. After incubation for 2 weeks, the cell colonies were fixed and stained by crystal violet (Sigma-Aldrich, Steinheim, Germany) and colonies of more than 50 cells were counted and analyzed.
Apoptosis analysis
Exponentially growing cells seeded in six-well plates were treated with (S)-crizotinib at different concentrations or 10 µl of DMSO as a control. Cell apoptosis was analyzed 3 days later by staining with annexin V and propidium iodide (PI) (KeyGEN BioTECH, Nanjing, China) according to the manufacturer’s instructions. After incubation for 5 min at room temperature in the dark, the apoptosis levels were analyzed using the flow cytometer (Becton Dickinson, Franklin Lakes, New Jersey, USA). Annexin V-positive and PI-negative/positive staining cells represent apoptotic cells.
Cell-cycle analysis
Exponentially growing cells seeded in six-well plates were treated with (S)-crizotinib at different concentrations or 10 µl of DMSO as controls. Three days after treatment, the cells were harvested and washed twice with cold PBS, and then fixed with cold 70% ethanol at 4°C overnight. To detect the DNA contents, the cells were stained with 50 μg/ml PI and 20 μg/ml RNase A (Beyotime Biotechnology) in the dark for 30 min at 37°C and analyzed using the flow cytometer.
Wound-healing assay
After incubation with free-serum medium for 12–24 h, cells at 90% confluence were scraped in two parallel lines with a P-10 pipette tip. The detached cells were washed off twice gently, and the medium was then replaced with 1% FBS complete medium containing (S)-crizotinib at different concentrations or DMSO. Images were taken at 0 and 24 h after scraping, respectively. The percentage of wound closure (original width−width of 24 h after scraping/original width) was calculated.
Transwell migration assay
In-vitro cell migration was assayed in a Transwell chamber (Corning Costar, Rochester, New York, USA) containing a gelatin-coated polycarbonate membrane filter (pore size: 8 μm). A total of 3×104 cells in 100 µl serum-free medium containing 5 µmol/l (S)-crizotinib or DMSO were seeded into the upper part of each chamber. Culture medium with 20% FBS was added to the lower chamber to stimulate cell migration. After 24-h incubation at 37°C with 5% CO2, the cells were stained with crystal violet (Sigma-Aldrich, Munich, Germany). The numbers of cells on the underside of the filters were observed and calculated by counting five random fields under a microscope (Jiangnan XD-202; Nanjing Jiangnan Novel Optics Co., Ltd, Jiangsu, China) at a magnification of ×200.
8-Oxo-dG assay
8-Oxo-dG levels were measured by Cy3-conjugated avidin, which has showed a high specificity to 8-oxodG previously 30. A total of 2×104 cells were seeded on each round coverslip in 24-well plates the day before treatment with (S)-crizotinib or DMSO. Three days after treatment, cells were fixed in pure methanol for 20 min and thereafter incubated for 15 min in TBS with 0.1% Triton X-100. Blocking was performed in 15% FBS and 0.1% Triton X-100 in TBS for 2 h at room temperature. Cells were then incubated with Cy3-conjugated avidin (1 μg/ml) (Rockland Immunochemicals, Limerick, Pennsylvania, USA) in blocking solution for 1 h at 37°C. After washing in PBS three times for 5 min each time, nuclei were stained with 4′,6-diamidino-2-phenylindole (Beyotime Biotechnology). After washing in PBS three times for 5 min each time, coverslips were transferred onto glass slides and images were acquired by a confocal microscope (Zeiss, Oberkochen, Germany).
Measurement of intracellular oxygen reactive species
Intracellular ROS levels were measured by flow cytometry using a cell-based ROS assay kit (Beyotime Biotechnology). Cells grown in six-well plates were treated with DMSO, 5 μmol/l (S)-crizotinib alone, or combined with 5 mmol/l NAC for 24 h, and then cells were harvested and washed twice with PBS. After incubation with 10 μmol/l dichlorofluorescein diacetate for 30 min at 37°C, the cells were analyzed using the FACSCaliber flow cytometer (Becton Dickinson). Intracellular ROS levels were expressed as the average dichlorodihydrofluorescein fluorescence intensity of the cells.
Statistical analysis
All experiments were repeated at least three times. Data were expressed as mean±SD. Statistical analysis was carried out using GraphPad Prism (GraphPad Software, La Jolla, California, USA). Data were analyzed using Student’s t-test or one-way analysis of variance where appropriate. χ2-Test was used to analyze the relationships between the results of IHC and clinical pathological features. IC50 of (S)-crizotinib was calculated using GraphPad Prism. P values less than 0.05 were considered significant.
Results
MTH1 is highly expressed in osteosarcoma tissues
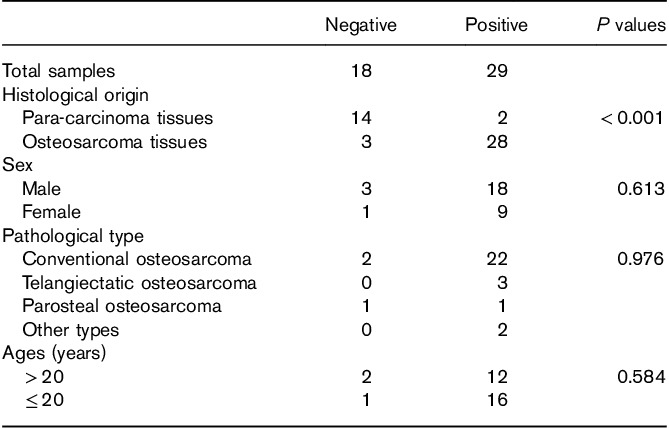
We used IHC to detect the expression of MTH1 in OS tissues and analyze its correlation with the clinical prognosis. Of the 31 OS patients, 28 (90.3%) showed positive MTH1 expression (Fig. 1 a and b) and three (9.7%) showed negative MTH1 expression (Fig. 1c). In the 16 corresponding adjacent normal tissue samples, only two (12.5%) showed positive MTH1 expression. The difference in MTH1 expression between OS and the corresponding adjacent normal tissues was found to be statistically significant (P<0.001). However, there was no significant correlation between MTH1 expression and sex, age, and pathological type (P>0.05). The results are shown in Table 1.
Fig. 1.
Expression of MTH1 in osteosarcoma and the corresponding adjacent tissues detected by immunohistochemical staining (magnification, ×200). (a, b) Positive expression of MTH1 in osteosarcoma tissues. (c) Negative expression of MTH1 in osteosarcoma tissues. (d) Negative expression of MTH1 in the corresponding adjacent tissues. The arrows in (a, b) represent positive expression and the arrows in (c, d) represent negative expression.
MTH1 is highly expressed in osteosarcoma cell lines and plays an important role in their proliferation
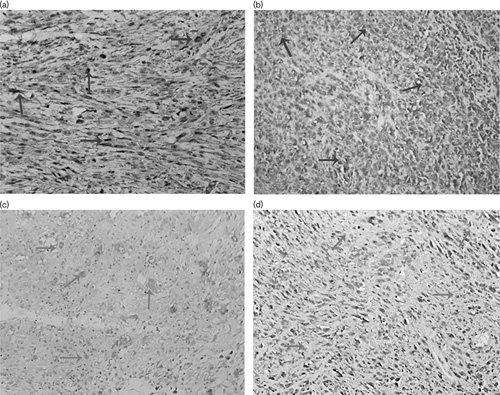
We used western-blotting analyses to detect the expression of MTH1 in OS cells and normal osteoblastic cell line. The results (Fig. 2a) indicated that MTH1 was highly expressed in OS cell lines but slightly expressed in the normal osteoblastic cell line hFOB, which indicated that the survival of OS cells, but not normal osteoblastic cells, was dependent on MTH1. To confirm this speculation, we used the CCK8 assay to explore the effects of MTH1 knockdown on the proliferation of OS cell lines. The results showed that the proliferations of MNNG/HOS, U2OS, and MG63 were significantly inhibited after knockdown of MTH1 with siRNA technology (Fig. 2d, P<0.001 compared with the NT group). Therefore, we could conclude that MTH1 plays an important role in the proliferation of OS cell lines.
Fig. 2.
MTH1 is highly expressed in osteosarcoma cell lines and plays an important role in their proliferation. (a) The expression of MTH1 in osteosarcoma cell lines and normal osteoblastic cell line hFOB detected by western blot. (b) Proteins extracted and analyzed with western blot 48 h after transfection. MTH1 protein was significantly decreased in the MTH1 siRNA group compared with the NT group. (c) Relative MTH1 gene expression analyzed with RT-PCR 48 h after transfection. **P<0.01, Student’s t-test. (d) U2OS, MNNG/HOS, and MG63 cells were transfected with MTH1 siRNA or nontargeting siRNA (NT) 3 days after transfection cell viability was detected. Data are shown as mean±SD from three independent experiments. ***P<0.001, Student’s t-test.
The sensitivity of osteosarcoma cell lines to (S)-crizotinib
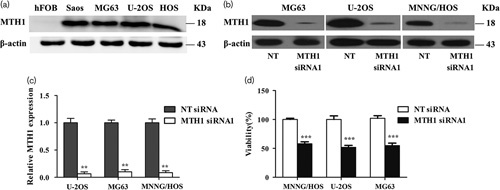
Several MTH1-targeting inhibitors have been explored in preclinical researches 23,24,27,28. Huber et al. 23 recently reported a potent MTH1 inhibitor (S)-crizotinib by screening a kinase inhibitor collection in a thermal shift stability assay. They found that (S)-crizotinib was attractive and interesting because it differs from a safe and marketed drug (R)-crizotinib only in one chiral center, which makes it more likely to have drug-like properties and potential clinical application values. Therefore, we choose (S)-crizotinib to explore the anticancer effects on OS cells by inhibiting MTH1. After treatment with (S)-crizotinib for 3 days, cell viability of U2OS, MNNG/HOS, and MG63 cells was detected using the CCK8 assay. The results showed that the IC50 value of (S)-crizotinib in U2OS, MG63, and MNNG/HOS was 4.967, 5.371, and 4.822 μmol/l, respectively (Fig. 3a). According to the IC50 value of each cell line, we found that there was no obvious difference in the sensitivity of three cell lines to (S)-crizotinib. Therefore, we randomly selected MG63 and MNNG/HOS to further study other anticancer effects of (S)-crizotinib on OS cells.
Fig. 3.
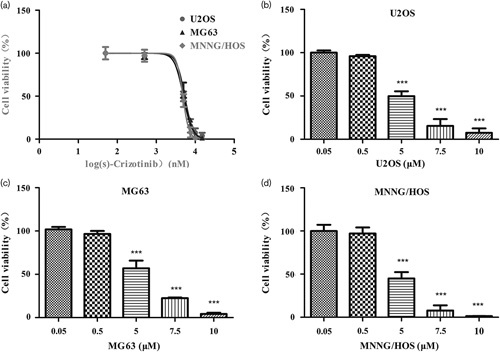
Dose–response curves of osteosarcoma cell lines inhibited by (S)-crizotinib from the CCK8 assay. Cells were treated with DMSO, 0.05, 0.5, 5, 7.5, 10, and 15 μmol/l (S)-crizotinib for 3 days, after which cell viability was determined using the CCK8 assay. (a) (S)-Crizotinib inhibited MTH1 with an IC50 value of 4.967, 5.371, and 4.822 μmol/l in U2OS, MG63, and MNNG/HOS, respectively. Data are shown as mean±SD from three independent experiments. Dose–response curves were calculated by GraphPad Prism. (b–d) Images are representative of the viability of U2OS, MG63, and MNNG/HOS. Data are shown as mean±SD from three independent experiments. ***P<0.001 compared with the 0.05 μmol/l group, one-way analysis of variance, followed by the least significant difference test.
(S)-Crizotinib inhibited cell proliferation and induced cell-cycle arrest and increased the rate of apoptosis in osteosarcoma cells
According to the dose–response curves and IC50 values of OS cell lines, we used 2.5 and 5 μmol/l (S)-crizotinib to represent the low-dose and the high-dose group, respectively. From the results of the colony formation assay (Fig. 4a), we can conclude that cell proliferation of OS cells was inhibited by (S)-crizotinib in a dose-dependent manner. To explore the potential mechanisms of the inhibitory effects of (S)-crizotinib, we used PI staining to study the distribution of cell cycle and annexin V and PI double staining to detect the apoptosis rate of OS cells. After treatment with 5 μmol/l (S)-crizotinib, cells in the G0/G1 phase were significantly increased and cells in the S phase were significantly decreased compared with the DMSO group (Fig. 4b and c, P<0.05 in MNNG/HOS and P<0.01 in MG63). The results of apoptosis analysis showed that the rates of apoptosis in the high-dose group were significantly increased (Fig. 4d and e, P<0.05 compared with the DMSO group). Thus, (S)-crizotinib inhibition of OS cell proliferations appears to be partially mediated by inducing G0/G1 phase arrest and apoptosis.
Fig. 4.
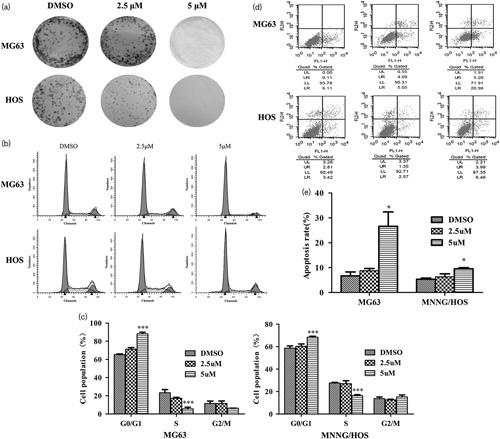
(S)-Crizotinib inhibited osteosarcoma cells proliferation partially by inducing cell-cycle arrest and increasing the rate of apoptosis. (a) (S)-Crizotinib inhibited osteosarcoma cells’ colony formation. Cells were treated with DMSO, 2.5, and 5 μmol/l (S)-crizotinib for 2 weeks, after which cells were fixed and stained with crystal violet. Images were obtained from three independent experiments. (b, c) Cell-cycle analysis by propidium iodide (PI) staining: cells were treated with DMSO, 2.5, and 5 μmol/l (S)-crizotinib for 3 days. Data are shown as mean±SD from three independent experiments. ***P<0.001 compared with the DMSO group, one-way analysis of variance, followed by the least significant difference test. (d, e) Flow cytometric analysis of apoptotic cells by annexin V and PI double staining. Data are shown as mean±SD from three independent experiments. *P<0.05 compared with the DMSO group, one-way analysis of variance, followed by the least significant difference test.
(S)-Crizotinib inhibited the migratory activity of osteosarcoma cells
To assess the effect of (S)-crizotinib on the migratory capability, we performed wound healing and a transwell chamber assay in MG63 and MNNG/HOS cells. As shown in Fig. 5a and b, the transwell assay showed that the number of migratory cells in the high-dose group was significantly decreased compared with that in the low-dose group. In addition, after treatment with 5 μmol/l (S)-crizotinib, the ability of OS cells to close a gap was significantly inhibited in the wound-healing assay (Fig. 5c and d). These results implied that MTH1 is involved in the mechanisms relevant to the promotion of OS cell migration.
Fig. 5.
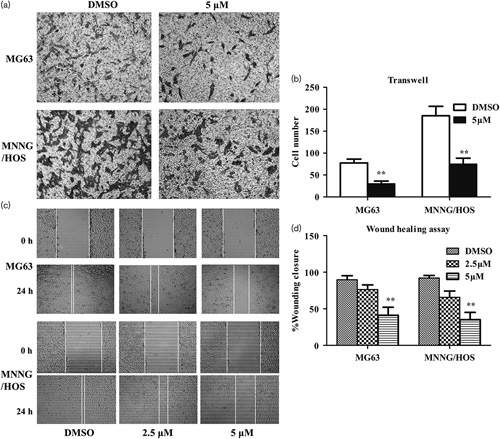
(S)-Crizotinib inhibited the migratory activities of osteosarcoma cells. (a) Transwell assay of osteosarcoma cells treated with DMSO or 5 μmol/l (S)-crizotinib: images were taken at magnification ×100. Photos are representative fields of cells penetrating the membrane and fixed in pure methanol, stained with crystal violet, and counted. the membrane. (b) Quantitative analysis of the transwell assay. Bars are means±SD from three independent experiments. **P<0.01 compared with the DMSO group, Student’s t-test. (c) Wound-healing assay of osteosarcoma cells treated with DMSO, 2.5, or 5 μmol/l (S)-crizotinib: images were taken at magnification ×40. (d) Quantitative analysis of the wound-healing assay. The percentage of wound closure=original width–width of 24 h after cell migration/original width. Bars are mean±SD from three independent experiments. **P<0.01 compared with the DMSO group, one-way analysis of variance, followed by the least significant difference test.
The effects of (S)-crizotinib on osteosarcoma were produced through targeting MTH1 and activating reactive oxygen species
Avidin has been shown to bind to 8-oxo-dGTP with high specificity 30. Therefore, to confirm that the effects of (S)-crizotinib on OS cells were produced through inhibiting MTH1, we used immunofluorescent staining with Cy3-conjugated avidin to detect the intracellular 8-oxo-dGTP levels in OS cell lines before and after (S)-crizotinib treatment. The results showed that the fluorescence intensity was significantly increased in MG63 and MNNG/HOS cell lines 72 h after treatment with 5 μmol/l (S)-crizotinib (Fig. 6a–c). These data indicated that compared with the DMSO group, (S)-crizotinib can statistically significantly increase the intracellular 8-oxo-dGTP levels, which indicates increased DNA oxidative damage levels in OS cells.
Fig. 6.
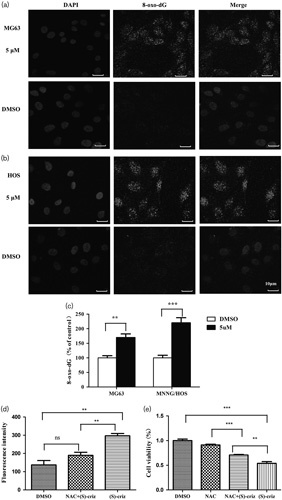
The effects of (S)-crizotinib on osteosarcoma were produced through targeting MTH1 and activating ROS. (a, b) Detection of 8-oxod-GTP incorporation into DNA in MG63 and MNNG/HOS cells treated with DMSO, 5 μmol/l (S)-crizotinib by staining with Cy3-conjugated avidin. (c) Quantification of 8-oxod-GTP incorporation into DNA: bars are mean±SD from three independent experiments. **P<0.01 and ***P<0.001 compared with the DMSO group, Student’s t-test. (d) Quantification of data of intracellular reactive oxygen species by dichlorofluorescin fluorescence in OS cells following exposure to DMSO, 5 μmol/l (S)-crizotinib alone, or combined with 5 mmol/l NAC for 24 h. Bars are mean±SD from three independent experiments. P>0.05, **P<0.01, Student’s t-test. (e) Cell viability determined by the CCK8 assay in OS cells following exposure to DMSO, 5 mmol/l NAC, 5 μmol/l (S)-crizotinib alone, or combined with 5 mmol/l NAC for 72 h. Bars are mean±SD from three independent experiments. **P<0.01 and ***P<0.001, Student’s t-test.
To determine whether (S)-crizotinib could induce ROS and its effect on OS cells, first, we detected the ROS level after treatment with DMSO, 5 μmol/l (S)-crizotinib alone, or combined with 5 mmol/l anti-oxidant NAC. The results implied that intracellular ROS was significantly increased following (S)-crizotinib exposure (Fig. 6d, P<0.01 compared with the DMSO group and the NAC+(S)-crizotinib group in MG63). To counteract the effects of ROS-induced (S)-crizotinib, we added NAC to (S)-crizotinib-treated cells. The results indicated that the cell viability was obviously inhibited by treatment with (S)-crizotinib alone or combined with NAC (Fig. 6e, P<0.001 for the (S)-crizotinib group compared with the DMSO group and P<0.01 for the (S)-crizotinib+NAC group compared with the NAC group). Thus, NAC could not completely attenuate the effect of (S)-crizotinib on OS cells. Taking the increased intracellular 8-oxo-dGTP levels together, we can deduce that the effects of (S)-crizotinib on OS were produced partially through inhibition of the MTH1 enzyme. Further, we found that there was also a significant difference in cell viability between the (S)-crizotinib group and the NAC+(S)-crizotinib group (Fig. 6e, P<0.01), which indicates that ROS induced by (S)-crizotinib also plays a very important role in the anticancer effects of (S)-crizotinib. Thus, we can conclude that the effects of (S)-crizotinib on OS were produced through targeting MTH1 and activating ROS.
Discussion
Anticancer drugs targeting the DDR pathway have become new rising stars and obtained huge success in breast cancer; the best example is the treatment of BRCA1/2-mutated patients with PARP1 inhibitors 31–33. The concept of synthetic lethality is an approach to target a normally nonessential pathway that becomes essential for cancer survival only in the context of a pre-existing gene mutation. This concept makes DDR targeted drugs more potent and widely used. Recently, ‘cancer phenotypic lethality’ has been proposed to describe a treatment that targets a nonessential enzyme in normal cells, but generally required for the survival of the cancer phenotype 34. This treatment may be advantageous to targeting the genotype as it can tackle the problem of intratumor heterogeneity independent of the genotype in cancer 5,24. Another obvious benefit is that targeting nonessential enzyme can spare normal cells while preferentially killing cancer cells and minimizing acute side effects, thus allowing such drugs to be administered as a maintenance therapy in many indications 34. There have not been potent and widely used targeting drugs for OS, which is in part because of the considerable heterogeneity of genetic alterations 35. Therefore, from the above, perhaps ‘cancer phenotypic lethality’ could represent a novel and promising therapeutic strategy for OS in the future. Here, targeting MTH1 is a good example of ‘cancer phenotypic lethality’.
The normally nonessential enzyme MTH1 is crucial for the repair of DNA oxidative damage, and it also plays an important role in the genesis and development of RAS-positive cancers 22,36,37. MTH1 has been reported to be highly expressed in various cancers 15–21. For the abnormally high intracellular ROS level in OS cells 25,26, we speculate that MTH1 may be crucial for the repair of DNA oxidative damage in OS. From the results of IHC, we found that MTH1 was positively expressed in 28 (90.32%) of 31 patients. The difference in MTH1 expression between OS and corresponding adjacent normal tissues was found to be statistically significant (P<0.001). However, other clinicopathological parameters may have no relationship with MTH1 expression, but these conclusions should be confirmed further because of the limited clinical data. From the results of western-blotting analyses, we can also find that the MTH1 expression of OS cells is significantly higher than that of the osteoblastic cell line. To clarify the effect of MTH1 on the survivability of OS cells, we used siRNA technology to decrease MTH1 expression and the results from CCK8 showed that the proliferation of OS was significantly inhibited after knockdown of MTH1, which confirmed that MTH1 played an important role in the survival of OS.
Several MTH1 inhibitors have been explored in preclinical studies 23,24,27,28. Among these inhibitors, (S)-crizotinib, the (S)-enantiomer of crizotinib, has greater potential to be used in clinic because of its pharmacokinetic and pharmacodynamic properties 23. To further explore the use of MTH1 as a therapeutic target for OS, we used MTH1-targeted drug (S)-crizotinib to investigate its anticancer effect on OS cell lines by in-vitro assays. From the dose–response curves of OS cell lines after treatment with (S)-crizotinib, we found that the IC50 value of (S)-crizotinib in U2OS, MG63, and MNNG/HOS was 4.967, 5.371, and 4.822 μmol/l, respectively. There was no obvious difference in the sensitivity of three cell lines to (S)-crizotinib and the IC50 value is similar to that in other researches 23. Next, we found that the proliferation of OS cells was significantly inhibited by (S)-crizotinib in a dose-dependent manner. Inhibition of MTH1 could theoretically increase intracellular 8-oxo-dGTP levels, which would cause DNA mutations and DNA strand breakage. To provide sufficient time to deal with these unrepaired DNA damages, cells would be regulated into a cell-cycle arrest. If these damages cannot be correctly repaired, cell death occurs 3–5,28. Consistent with these mechanisms, the results from flow cytometry showed that (S)-crizotinib could induce cell-cycle arrest in the G0/G1 phase and triggered cell apoptosis in OS cells, which indicated that the MTH1-targeted drug (S)-crizotinib may regulate osteosarcoma development and growth by affecting cell-cycle progression and apoptosis.
OS migratory ability is essential for the initial metastatic or recurrent process, and the rapid and frequent development of lung metastasis is the major cause of death in patients with OS 1. Thus, therapies inhibiting the migratory ability of osteosarcoma cells may decrease local recurrence or metastasis, thereby improving survival. Interestingly, the results of wound healing and the transwell migration assay showed that (S)-crizotinib could inhibit the migratory ability of OS cells, which was the first time to be reported as far as we know. However, the specific mechanism is still unknown and needs to be further studied.
Huber et al. 23 found that MTH1 is the most potent and specific target for (S)-crizotinib, whereas it showed no obvious inhibiting effects on kinases. The intracellular 8-oxo-dGTP level would be significantly increased after inhibition of MTH1 with (S)-crizotinib. Avidin has been shown to bind to 8-oxo-dGTP with high specificity 30. Therefore, we used immunofluorescent staining with Cy3-conjugated avidin to detect the intracellular 8-oxo-dGTP levels in OS cell lines before and after (S)-crizotinib treatment. The results showed that compared with the DMSO group, (S)-crizotinib can statistically significantly increase the intracellular 8-oxo-dGTP levels, which indicates increased DNA oxidative damage levels in OS cells. Recently, (S)-crizotinib was also shown to induce ROS in human NSCLC cells 29. To explore whether (S)-crizotinib could induce ROS and its effect on osteosarcoma cells, first, we detected the ROS level after treatment with (S)-crizotinib. The results implied that (S)-crizotinib could significantly increase intracellular ROS levels in OS cells. To counteract the effects of ROS induced by (S)-crizotinib, we added NAC to (S)-crizotinib treating cells. The results indicated that the cell viability was obviously inhibited by treatment with not only (S)-crizotinib alone but also (S)-crizotinib combined with NAC. Thus, NAC could not completely attenuate the effect of (S)-crizotinib on OS cells. Taking the increased intracellular 8-oxo-dGTP level and significantly inhibited proliferation of OS cells after knockdown of MTH1 together, we can conclude that the effects of (S)-crizotinib on OS were produced partially through inhibition of MTH1 enzyme. Further, we found that there was also a significant difference in cell viability between the (S)-crizotinib group and the NAC+(S)-crizotinib group, which indicates that ROS induced by (S)-crizotinib also plays a very important role in the anticancer effects of (S)-crizotinib. Therefore, we can conclude that the effects of (S)-crizotinib on osteosarcoma were produced through targeting MTH1 and activating ROS.
Conclusion
MTH1 was found to be highly expressed in OS tissues and cell lines. The viability of osteosarcoma cells was significantly decreased after MTH1 knockdown with siRNA technology. Furthermore, through in-vitro studies, we confirmed that (S)-crizotinib could inhibit the proliferation of OS cells by increasing the apoptosis levels and causing G0/G1 arrest by targeting MTH1 and activating ROS. In addition, (S)-crizotinib could inhibit the migration of OS cells. This study will provide a promising therapeutic target and the theoretical basis for the clinical application of (S)-crizotinib in OS. However, these anticancer effects of (S)-crizotinib need to be tested further by in-vivo experiments. Moreover, the effects of inhibiting MTH1 will be much more intensive after the ROS levels are increased 27. Among all these first-line chemotherapy drugs for OS, doxorubicin could increase ROS levels in OS cells 38. Thus, in our future researches, we will focus on the synergistic anticancer effect of (S)-crizotinib combined with doxorubicin.
Acknowledgements
The authors thank Yunlu Liu for providing the U2OS cell line, which was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and for providing technical help.
Authors’ contributions: Xiangcheng Qing made contributions to research design, the acquisition, analysis or interpretation of data, and drafting of the paper and revising it critically; Xiao Lv made contributions to analysis or interpretation of data and drafting of the paper or revising it critically; Zengwu Shao, the corresponding author, made contributions to research design, revising the paper, and approval of the submitted and final versions. Deyao Shi made contributions to revising the manuscript and conducting additional experiments suggested by reviewers. All of the other authors took part in the research design. All authors have read and approved the final submitted manuscript.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Jaffe N. Osteosarcoma: review of the past, impact on the future. The american experience. Cancer Treat Res 2009; 152:239–262. [DOI] [PubMed] [Google Scholar]
- 2.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. Parp inhibition: Parp1 and beyond. Nat Rev Cancer 2010; 10:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 6.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010; 44:479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magder S. Reactive oxygen species: Toxic molecules or spark of life? Crit Care 2006; 10:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schieber M, Chandel NS. Ros function in redox signaling and oxidative stress. Curr Biol 2014; 24:R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichikawa J, Tsuchimoto D, Oka S, Ohno M, Furuichi M, Sakumi K, Nakabeppu Y. Oxidation of mitochondrial deoxynucleotide pools by exposure to sodium nitroprusside induces cell death. DNA Repair (Amst) 2008; 7:418–430. [DOI] [PubMed] [Google Scholar]
- 10.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes g----t and a----c substitutions. J Biol Chem 1992; 267:166–172. [PubMed] [Google Scholar]
- 11.Leiros I, Nabong MP, Grosvik K, Ringvoll J, Haugland GT, Uldal L, et al. Structural basis for enzymatic excision of n1-methyladenine and n3-methylcytosine from DNA. EMBO J 2007; 26:2206–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowerman B. Cell biology. Oxidative stress and cancer: a beta-catenin convergence. Science 2005; 308:1119–1120. [DOI] [PubMed] [Google Scholar]
- 13.Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial dnas. EMBO J 2008; 27:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cdna for a human enzyme that hydrolyzes 8-oxo-dgtp, a mutagenic substrate for DNA synthesis. J Biol Chem 1993; 268:23524–23530. [PubMed] [Google Scholar]
- 15.Borrego S, Vazquez A, Dasi F, Cerda C, Iradi A, Tormos C, et al. Oxidative stress and DNA damage in human gastric carcinoma: 8-oxo-7′8-dihydro-2′-deoxyguanosine (8-oxo-dg) as a possible tumor marker. Int J Mol Sci 2013; 14:3467–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coskun E, Jaruga P, Jemth AS, Loseva O, Scanlan LD, Tona A, et al. Addiction to mth1 protein results in intense expression in human breast cancer tissue as measured by liquid chromatography–isotope-dilution tandem mass spectrometry. DNA Repair (Amst) 2015; 33:101–110. [DOI] [PubMed] [Google Scholar]
- 17.Eshtad S, Mavajian Z, Rudd SG, Visnes T, Bostrom J, Altun M, Helleday T. Hmyh and hmth1 cooperate for survival in mismatch repair defective t-cell acute lymphoblastic leukemia. Oncogenesis 2016; 5:e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obtulowicz T, Swoboda M, Speina E, Gackowski D, Rozalski R, Siomek A, et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis 2010; 25:463–471. [DOI] [PubMed] [Google Scholar]
- 19.Okochi E, Watanabe N, Sugimura T, Ushijima T. Single nucleotide instability: A wide involvement in human and rat mammary carcinogenesis? Mutat Res 2002; 506-507:101–111. [DOI] [PubMed] [Google Scholar]
- 20.Song WJ, Jiang P, Cai JP, Zheng ZQ. Expression of cytoplasmic 8-oxo-gsn and mth1 correlates with pathological grading in human gastric cancer. Asian Pac J Cancer Prev 2015; 16:6335–6338. [DOI] [PubMed] [Google Scholar]
- 21.Speina E, Arczewska KD, Gackowski D, Zielinska M, Siomek A, Kowalewski J, et al. Contribution of hmth1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J Natl Cancer Inst 2005; 97:384–395. [DOI] [PubMed] [Google Scholar]
- 22.Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Strom CE, et al. Mth1 inhibition eradicates cancer by preventing sanitation of the dntp pool. Nature 2014; 508:215–221. [DOI] [PubMed] [Google Scholar]
- 23.Huber KV, Salah E, Radic B, Gridling M, Elkins JM, Stukalov A, et al. Stereospecific targeting of mth1 by (s)-crizotinib as an anticancer strategy. Nature 2014; 508:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleh A, Gokturk C, Warpman-Berglund U, Helleday T, Granelli I. Development and validation of method for th588 and th287, potent mth1 inhibitors and new anti-cancer agents, for pharmacokinetic studies in mice plasma. J Pharm Biomed Anal 2015; 104:1–11. [DOI] [PubMed] [Google Scholar]
- 25.Hu R, Guille M, Arbault S, Lin CJ, Amatore C. In situ electrochemical monitoring of reactive oxygen and nitrogen species released by single mg63 osteosarcoma cell submitted to a mechanical stress. Phys Chem Chem Phys 2010; 12:10048–10054. [DOI] [PubMed] [Google Scholar]
- 26.Nathan FM, Singh VA, Dhanoa A, Palanisamy UD. Oxidative stress and antioxidant status in primary bone and soft tissue sarcoma. BMC Cancer 2011; 11:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warpman Berglund U, Sanjiv K, Gad H, Kalderen C, Koolmeister T, Pham T, et al. Validation and development of mth1 inhibitors for treatment of cancer. Ann Oncol 2016; 27:2275–2283. [DOI] [PubMed] [Google Scholar]
- 28.Dong L, Wang H, Niu J, Zou M, Wu N, Yu D, et al. Echinacoside induces apoptotic cancer cell death by inhibiting the nucleotide pool sanitizing enzyme mth1. Onco Targets Ther 2015; 8:3649–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai X, Guo G, Zou P, Cui R, Chen W, Chen X, et al. (S)-crizotinib induces apoptosis in human non-small cell lung cancer cells by activating ROS independent of MTH1. J Exp Clin Cancer Res 2017; 36:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struthers L, Patel R, Clark J, Thomas S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal Biochem 1998; 255:20–31. [DOI] [PubMed] [Google Scholar]
- 31.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of brca2-deficient tumours with inhibitors of poly(adp-ribose) polymerase. Nature 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 32.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in brca mutant cells as a therapeutic strategy. Nature 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 33.Helleday T. The underlying mechanism for the parp and brca synthetic lethality: Clearing up the misunderstandings. Mol Oncol 2011; 5:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helleday T. Cancer phenotypic lethality, exemplified by the non-essential mth1 enzyme being required for cancer survival. Ann Oncol 2014; 25:1253–1255. [DOI] [PubMed] [Google Scholar]
- 35.Bousquet M, Noirot C, Accadbled F, Sales de Gauzy J, Castex MP, Brousset P, Gomez-Brouchet A. Whole-exome sequencing in osteosarcoma reveals important heterogeneity of genetic alterations. Ann Oncol 2016; 27:738–744. [DOI] [PubMed] [Google Scholar]
- 36.Patel A, Burton DG, Halvorsen K, Balkan W, Reiner T, Perez-Stable C, et al. Mutt homolog 1 (mth1) maintains multiple kras-driven pro-malignant pathways. Oncogene 2015; 34:2586–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai P, Young JJ, Burton DG, Giribaldi MG, Onder TT, Weinberg RA. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic ras-induced DNA damage and premature senescence. Oncogene 2011; 30:1489–1496. [DOI] [PubMed] [Google Scholar]
- 38.Shin SH, Choi YJ, Lee H, Kim HS, Seo SW. Oxidative stress induced by low-dose doxorubicin promotes the invasiveness of osteosarcoma cell line u2os in vitro. Tumour Biol 2016; 37:1591–1598. [DOI] [PubMed] [Google Scholar]


