Figure 1. Outline of the overall study design.
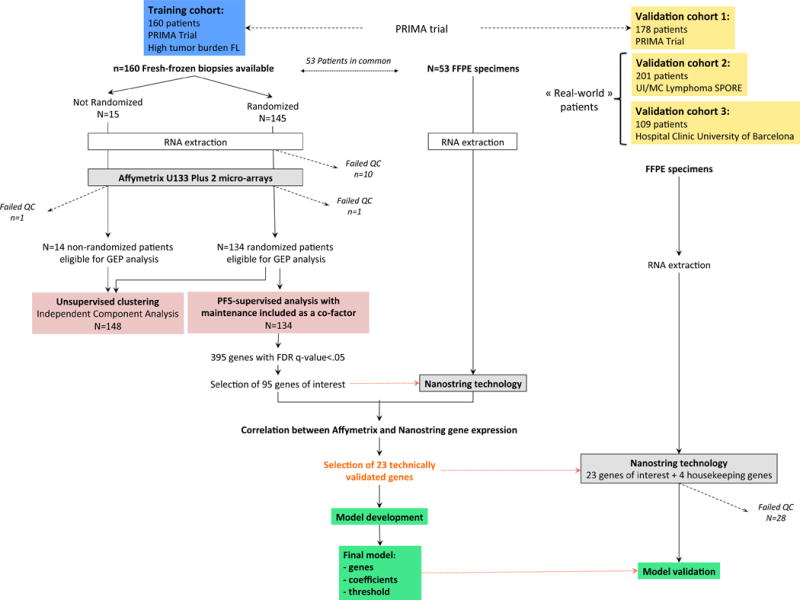
Fresh-frozen tissue (FFT) tumor biopsies were prospectively obtained from 160 untreated patients enrolled in the international PRIMA trial. RNA with sufficient quality (RIN>6.5) was obtained for 149/160 cases and gene-expression profiling was performed using Affymetrix U133 Plus 2.0 micro-arrays. A multivariate Cox regression analysis identified genes whose expression was associated with PFS independent of maintenance treatment in the subgroup of randomized patients. Expression levels from 95 curated genes were then determined by means of digital expression profiling (NanoString technology) in 53 FFPET samples of the training set, allowing assessment of the technical replication of expression levels for each gene between technologies. Genes with high correlation (>0.75) were included in a L2-penalized Cox model adjusted on rituximab maintenance to build a PFS-predictive score. The model was further evaluated using NanoString technology in 488 FFPET samples from 3 independent international cohorts of patients: a distinct validation set from the PRIMA trial (n=178), and two others obtained in large centers (respectively the Mayo Clinic/Iowa SPORE project, n=201 and the Barcelona Hospital Clinic, n=109). An unsupervised analysis of the gene-expression data generated in the training cohort was also performed independently.
Abbreviations: FFT: fresh-frozen tissues; FFPE: formalin-fixed paraffin-embedded tissues; PFS: progression-free survival.
