Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease with dismal response warranting the need for enhancing our understanding of AML biology. One prognostic feature associated with inferior response is the presence of activating mutations in FMS-like tyrosine kinase 3 (FLT3) especially occurrence of internal tandem duplication (FLT3-ITD). Although poorly understood, differential metabolic and signaling pathways associated with FLT3-ITD might contribute towards the observed poor prognosis. We performed a non-targeted global metabolic profiling of matched cell and plasma samples obtained at diagnosis to establish metabolic differences within FLT3-ITD and FLT3-WT pediatric AML. Metabolomic profiling by Ultra-High Performance-Liquid-Chromatography–Mass Spectrometry identified differential abundance of 21 known metabolites in plasma and 33 known metabolites in leukemic cells by FLT3 status. These metabolic features mapped to pathways of significant biological importance. Of interest were metabolites with roles in cancer, cell progression and involvement in purine metabolism and biosynthesis, cysteine/methionine metabolism, tryptophan metabolism, carnitine mediated fatty acid oxidation, and lysophospholipid metabolism. Although validation in a larger cohort is required, our results for the first time investigated global metabolic profile in FLT3-ITD AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by complex molecular and cytogenetic abnormalities. These abnormalities are used for risk stratification and prediction of clinical outcome in AML. In addition to cytogenetic lesions, a number of mutations of prognostic significance have been identified using next generation sequencing efforts in recent years. One of the genes of prognostic significance in AML is fms-like tyrosine kinase 3 (FLT3). FLT3 plays role in the regulation of cell proliferation, differentiation and survival1,2. Mutations in FLT3, especially internal tandem duplication (ITD) in the juxta-membrane domain (FLT3-ITD), are present in around 20% of AML patients with predominant occurrence in patients with cytogenetically normal AML. The presence of FLT3-ITD mutation results in constitutive activation of FLT3 signaling and has been associated with poor prognosis and clinical outcome in response to chemotherapy2–6 Tyrosine kinase inhibitors such as midostaurin, sunitinib, sorafenib and ibrutinib have been investigated for treatment of FLT3-ITD positive AML2,7. However, development of drug resistance to FLT3-inhibitors has been a major challenge in successful treatment of patients8,9, warranting the need for further understanding the complexity and biology of FLT3-ITD in AML.
The emerging field of metabolomics focuses on quantitating metabolic features to better understand biochemical alterations associated with cancer. In recent years, metabolic alterations of significant relevance have been identified in several solid tumors10,11 and hematological malignancies such as chronic lymphocytic leukemia12, childhood acute lymphoblastic leukemia13,14 and adult AML15,16. A distinct glucose metabolism signature and 2–hydroxyglutarate levels, which is also referred to as an ‘oncometabolite’ have been reported to be of prognostic significance in adult AML patients15,16 but there are no reports in pediatric AML. Among pediatric patients, differential metabolome in pediatric acute lymphoblastic leukemia patients as compared to healthy controls has been reported13. Overall, these studies show that metabolomics can be successfully applied in leukemia studies to improve our understanding of disease characteristics and variance in clinical outcomes in AML. Unfortunately, the metabolome of pediatric AML has not been studied and there is currently a gap in our understanding of any metabolic features that are specific to FLT3-ITD positive AML. Identifying the FLT3-ITD specific metabolic profile might enhance our understanding of the poor outcome in these patients as well as give insight of disease biology and drug resistance mechanism. Thus, the current study was designed with the goal to establish metabolomic differences between FLT3 wild-type (WT) and FLT3-ITD childhood AML patients.
Results
Patient cohort and metabolite profiling
Our study included matched plasma samples and leukemic cells obtained at diagnosis from 16 pediatric AML patients (FLT3-ITD n = 8 and FLT3-WT, n = 8) enrolled in the AAML1031 clinical trial (NCT01371981). Table 1 summarizes the patient characteristics; overall median age of the patients was 13 years. Plasma samples and matched leukemic cells were obtained primarily from peripheral blood samples from patients at diagnosis before initiation of chemotherapy.
Table 1.
Characteristics of pediatric patients with FLT3-ITD or FLT3-WT AML.
| Characteristics | FLT3-WT | FLT3-ITD |
|---|---|---|
| Total | 8 | 8 |
| Age (years) | ||
| Mean | 13.71 | 12.41 |
| Range | 6.89–19.42 | 4.16–17.79 |
| Sex | ||
| Male | 3 | 3 |
| Female | 5 | 5 |
| Race | ||
| White | 6 | 5 |
| Black | 1 | 0 |
| Other | 1 | 3 |
| Allelic Ratio | ||
| Average | 0 | 2.88 |
| Range | 0 | 0.8–13.35 |
| Cytogenetics | ||
| Normal | 3 | 6 |
| Abnormal | 5 | 2 |
Untargeted global metabolomics profiling was performed in both positive and negative ionization using Ultra-High performance Liquid Chromatography – Mass Spectrometry (UHPLC-MS) at Southeast Center for Integrated Metabolomics (SECIM) University of Florida. A total of 2966 features were identified in plasma specimens and 1742 features in leukemic cells. Using our internal metabolite library that is regularly updated by running standards, we were able to annotate 290 metabolites in plasma and 143 metabolites identified in cells to known metabolites of which 58 were common between both types of specimens.
Plasma Metabolic features differentiating FLT3-ITD and FLT3-WT AML
We utilized Metaboanalyst software17 to identify features separating FLT3-ITD from FLT3-WT AML. A total of 209 features were significantly different in abundance between FLT3-ITD and FLT3-WT groups at FDR <0.05 (volcano plot in Supplementary Figure 1 shows the separation of the two groups, Supplementary Table 1 provides list of metabolites). Among these 21 were annotated to known metabolites and were differentially abundant by FLT3 status as shown in Table 2. Metabolites with higher abundance in FLT3-ITD vs. FLT3-WT included: (1) organic acids: 3-methyl-2-oxovaleric acid associated with isoleucine metabolism, pyridine-2,3-dicarboxylate, 6-carboxyhexanoate (also commonly known as pimelic acid which has been reported to be higher in patients with uremic serum)18 and methyl indole-3-acetate; (2) amino acids and intermediates such as guanine, N-acetyl arginine, N-alpha-acetyl-L-Lysine, N-acetyl-DL-glutamic acid, L-carnitine, N-acetyl glycine, GABA, N-acetyl-amine, cysteine-S sulfate, and threonine/homoserine; (3) phosphocholine. Metabolites L-cysteic acid and asparagine were less abundant in FLT3-ITD vs. FLT3-WT patients.
Table 2.
Metabolites with significantly differential abundance in plasma samples from patients with FLT3-ITD vs. without FLT3-ITD.
| Metabolite | Classification | Associated Pathway | p-value | FDR | Fold Change |
|---|---|---|---|---|---|
| Guanine | Nucleosides | Purine metabolism/biosynthesis | 1.09E-08 | 8.01E-06 | 5.722 |
| Pyridine-2,3-Dicarboxylate | Organic Acids | Nicotinate and nicotinamide metabolism; beta-Alanine metabolism; tryptophan metabolism | 8.75E-07 | 1.98E-04 | 3.6429 |
| N-Alpha-Acetyl-L-Lysine | Amino Acids | Lysine synthesis | 1.36E-06 | 2.43E-04 | 6.0508 |
| N-Acetylglycine | Amino Acids | Arginine and proline metabolism | 2.33E-06 | 3.42E-04 | 3.6015 |
| GABA | Amino Acids | Alanine, aspartate and glutamate metabolism; Butanoate Metabolism; beta-Alanine Metabolism | 3.74E-06 | 4.78E-04 | 2.7014 |
| N-Acetyl-L-Alanine | Amino Acids | Arginine and proline metabolism | 8.90E-06 | 8.09E-04 | 2.5869 |
| Phosphocholine | Amines | Glycerophospholipid metabolism | 2.74E-05 | 1.78E-03 | 1.8098 |
| Diphenylamine | Xenobiotics | N/A | 3.54E-05 | 2.17E-03 | 2.0411 |
| 3-Methyl-2-Oxovaleric Acid | Organic Acids | Isoleucine Metabolism | 1.13E-04 | 4.83E-03 | 43.236 |
| L-Carnitine | Amino Acids | Fatty Acid Metabolism | 1.74E-04 | 6.16E-03 | 1.9491 |
| Cysteine-S-sulfate | Amino Acids | Cysteine and methionine metabolism | 2.27E-04 | 7.03E-03 | 3.7072 |
| 6-Carboxyhexanoate | Organic Acids | 2.76E-04 | 8.20E-03 | 8.1077 | |
| Methyl Indole-3-Acetate | Organic Acids | Tryptophan metabolism | 4.70E-04 | 0.01191 | 1.8294 |
| Threonine/Homoserine | Amino Acids | Aminoacyl-tRNA biosynthesis; Cysteine and methionine metabolism | 7.74E-04 | 0.017915 | 4.3347 |
| N-Acetyl-DL-Glutamic Acid | Amino Acids | Urea Cycle | 9.51E-04 | 0.019802 | 1.774 |
| N-Acetyl-Arginine | Amino Acids | Protein/Amino Acid biosynthesis | 1.27E-03 | 0.024796 | 11.915 |
| Asparagine | Amino Acids | Alanine, aspartate and glutamate metabolism; Nitrogen Metabolism; Cyanoamino acid Metabolism | 1.60E-03 | 0.030212 | 0.4477 |
| L-Cysteic Acid | Amino Acids | Taurine and hypotaurine metabolism; Cysteine and methionine metabolism | 2.27E-03 | 0.038341 | 0.23168 |
| 4-Acetamidobutanoate | Amino acids | N/A | 2.27E-03 | 0.038341 | 1.9979 |
| 3-Hydroxydecanoic acid | Organic Acids | N/A | 2.57E-03 | 0.041408 | 2.5453 |
| Betaine | Amino acids | Glycine, serine, and threonine metabolism | 2.59E-03 | 0.041654 | 1.6109 |
Note: Metabolites with lower abundance in FLT3-ITD are in italics.
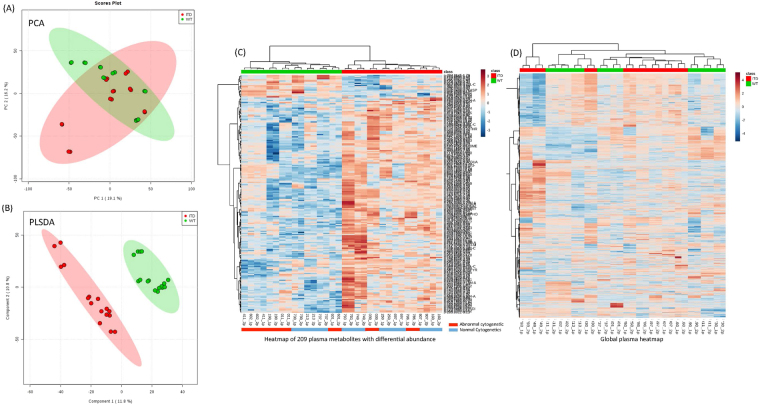
Multivariate analyses were conducted to evaluate the effectiveness of discriminating metabolomes according to FLT3 status. The principal component analysis (PCA) plots for an unbiased multivariate analysis did not clearly separate by FLT3 groups (Fig. 1A); however, Partial Least Square- Discriminant Analysis (PLSDA), a supervised method of multivariate analysis for discrimination in-group modeling, showed clear discrimination between FLT3-WT and FLT3-ITD (Fig. 1B). Permutation tests were conducted to identify overfitting due to supervision in the model and thresholds were set at Q2 > 0.2. The Q2 score was 0.53 in plasma metabolome model, indicating classification of two groups. Figure 1C shows heatmap of the 209 plasma metabolites (annotated as well as un-annotated) with significantly different abundance between FLT3-ITD and FLT3-WT patients (additionally, Fig. 2D shows heatmap of all the metabolites detected in plasma samples).
Figure 1.
Plasma Metabolic differences between FLT3-ITD and FLT3-WT AML. (A) PCA plots of plasma samples. Unbiased multivariate analysis does not show clear separation of samples based on FLT3 status groups. (B) PLSDA plot of plasma samples shows global separation of pediatric AML patients by FLT3 status (FLT3-ITD n = 8 and FLT3-WT n = 8, all samples were run in duplicate). (C) Heatmap shows relative abundance patterns of 209 plasma metabolites (known and un-annotated) with significantly differential abundance according to FLT3 status. Clustering within the heatmap shows a clear distinction of several metabolites between in FLT-WT and FLT3-ITD patients. (D) Heatmap of the global metabolome for patient plasma samples from AML patients.
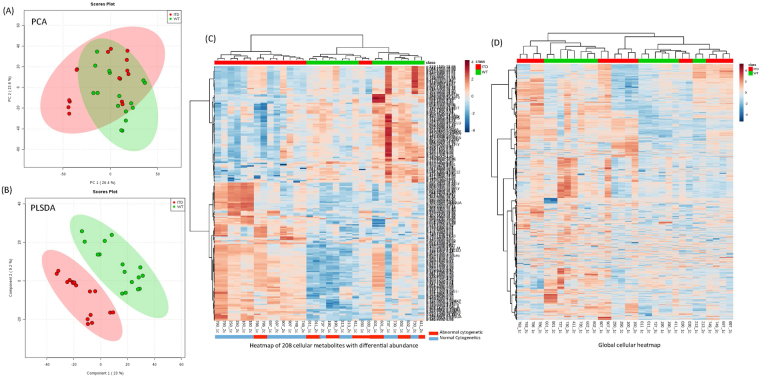
Figure 2.
Cellular Metabolic differences between FLT3-ITD and FLT3-WT AML. (A) PCA plots of cellular samples. Unbiased multivariate analysis does not show clear separation of samples based on FLT3 status groups. (B) PLSDA plot of shows global separation of pediatric AML patients by FLT3 status (FLT3-ITD n = 8 and FLT3-WT n = 8, all samples were run in duplicate). (C) Heatmap shows relative abundance patterns of 208 cellular metabolites (known and un-annotated) with significantly differential abundance according to FLT3 status. Clustering within the heatmap shows a clear distinction of several metabolites between in FLT-WT and FLT3-ITD patients. (D) Heatmap of the global metabolome for leukemic cells from AML patients.
Cellular Metabolic features differentiating FLT3-ITD and FLT3-WT AML
Evaluation of leukemic cells obtained at diagnosis between patients with FLT3-WT or FLT3-ITD status identified 208 (annotated and un-annotated) metabolites with differential abundance by FLT3 status in univariate analysis (p < 0.05; Supplementary Figure 2 shows the corresponding volcano plot and Supplementary Table 2 lists all the metabolites). Among the annotated metabolites within patient leukemic cell samples, as shown in Table 3, 33 were significantly different between FLT3-WT and FLT3-ITD groups. 16 metabolites had lower abundance and 17 had higher abundance in FLT3-ITD patients as compared to FLT3-WT group. Less abundant metabolites included tryptophan and formyl-5-hydroxykynurenamine with a role in tryptophan metabolism pathway; sugars such as disaccharide 6 C/6 C, glucose/fructose; glyceraldehyde with role in pyruvate metabolism and gluconeogenesis; fatty acids citramalate and arachidonic acid; and organic acids such as xylene sulfonate, benzoate, glycodeoxycholic acid and succinate (which plays a role in TCA cycle). Among metabolites with higher abundance in FLT3-ITD were: amino acids (leucine, L-carnitine, L-methionine, 4-oxoproline, LL-2,6-Diaminoheptanedioate and L-acetylcarnitine), nucleosides (guanosine, adenosine, inosine, hypoxanthine, adenosine 5′-monophosphate and guanine), lipids (Palmitoleic acid and Lyso PEs), an organic acid (glycolate), and one dipeptide (L-Leucyl-L-proline).
Table 3.
Metabolites with significantly differential abundance in leukemia cell samples from patients with FLT3-ITD vs. without FLT3-ITD.
| Metabolite | Classification | Associated Pathway | p-value | FDR | Fold Change |
|---|---|---|---|---|---|
| Xylenesulfonate | Organic Acids | N/A | 1.09E-06 | 0.0005 | 0.8515 |
| Succinate | Organic Acids | TCA Cycle; Alanine, aspartate, and glutamate metabolism; Propanoate Metabolism; Tyrosine Metabolism; Butanoate metabolism; Phenylalanine metabolism; Glyoxlyate and dicarboxylate metabolism | 1.61E-06 | 0.0005 | 0.8708 |
| Disaccharide-6C/6 C | Sugars | N/A | 1.79E-06 | 0.0005 | 0.7703 |
| L-Acetylcarnitine | Amino Acids | Fatty Acid Metabolism | 2.35E-06 | 0.0005 | 3.7810 |
| Glyceraldehyde/Lactate | Aldehydes | Glycolysis or Gluconeogenesis; Pyruvate Metabolism; Propanoate metabolism | 3.70E-06 | 0.0007 | 0.8751 |
| Glucose/Fructose | Sugars | Starch and sucrose metabolism; Galactose Metabolism; Pentose Phosphate Pathway; Amino sugar and nucleotide sugar metabolism | 5.37E-06 | 0.0008 | 0.8398 |
| Inosine | Nucleosides | Purine metabolism | 1.87E-05 | 0.0018 | 2.2740 |
| Adenosine 5′-Monophosphate | Nucleosides | Purine metabolism; Nitrogen metabolism | 3.40E-05 | 0.0028 | 3.0967 |
| Allopurinol | Xenobiotics | N/A | 4.26E-05 | 0.0032 | 2.0377 |
| Guanosine | Nucleosides | Purine metabolism/biosynthesis | 5.29E-05 | 0.0037 | 1.9649 |
| Hypoxanthine | Nucleosides | Purine metabolism/biosynthesis | 1.27E-04 | 0.0060 | 2.7834 |
| Adenosine | Nucleosides | Purine metabolism/biosynthesis | 1.36E-04 | 0.0061 | 2.1136 |
| LysoPE(p-526.2933–12.92; 22:6) | Lipids | Lysophospholipid Metabolism | 2.18E-04 | 0.0084 | 85.5160 |
| Tryptophan | Amino Acids | Tryptophan Metabolism, Nitrogen metabolism, Aminoacyl-tRNA metabolism | 6.26E-04 | 0.0184 | 0.9078 |
| LysoPE(n-500.2768-12.71; 20:4) | Lipids | Lysophospholipid Metabolism | 6.51E-04 | 0.0184 | 0.9160 |
| Benzoate | Organic Acids | N/A | 8.50E-04 | 0.0199 | 0.9075 |
| Formyl-5-hydroxykynurenamine | Organic Acids | Tryptophan Metabolism | 1.23E-03 | 0.0238 | 0.6343 |
| LysoPE(n-452.2772-13.35; 16:0) | Lipids | Lysophospholipid Metabolism | 1.28E-03 | 0.0238 | 0.9064 |
| C6H12O6-HEXOSE/KETOSE/INOSITOL | Sugars | Galactose Metabolism; Amino sugar and nucleotide sugar metabolism | 1.50E-03 | 0.0250 | 0.9202 |
| L-Carnitine | Amino Acids | Fatty Acid Metabolism | 1.59E-03 | 0.0260 | 1.9458 |
| LL-2,6-Diaminoheptanedioate | Amino Acids | Lysine Biosynthesis | 1.70E-03 | 0.0268 | 3.2397 |
| L-leucyl-L-proline | Dipeptides | N/A | 1.77E-03 | 0.0274 | 1.6180 |
| LysoPE(p-502.2908-12.90: 20:4) | Lipids | Lysophospholipid Metabolism | 1.94E-03 | 0.0284 | 20.0570 |
| Arachidonic Acid (20:4) | Fatty Acids | Arachidonic acid metabolism | 2.77E-03 | 0.0339 | 0.8431 |
| 4-oxoproline | Amino Acids | Arginine and proline metabolism | 3.63E-03 | 0.0382 | 2.1616 |
| Glycodeoxycholic acid | Organic Acids | N/A | 3.94E-03 | 0.0395 | 0.9135 |
| Palmitoleic acid | Lipids | Fatty Acid Biosynthesis | 4.07E-03 | 0.0400 | 1.6714 |
| Citramalate | Fatty Acids | N/A | 4.30E-03 | 0.0417 | 0.8111 |
| LysoPE(n-480.3097-14.59: 18:0) | Lipids | Lysophospholipid Metabolism | 5.05E-03 | 0.0452 | 0.8803 |
| Guanine | Nucleosides | Purine metabolism/biosynthesis | 5.27E-03 | 0.0466 | 3.3509 |
| L-Methionine | Amino Acids | Aminoacyl-tRNA biosynthesis; Cysteine and methionine metabolism | 5.71E-03 | 0.0484 | 2.0903 |
| Leucine | Amino Acids | Aminoacyl-tRNA biosynthesis; valine, leucine, and isoleucine biosynthesis and degradation | 5.76E-03 | 0.0484 | 1.9154 |
| 2-alpha-D-glucosyl-D-glucose | sugars/lipids | N/A | 5.78E-03 | 0.0484 | 0.9215 |
Note: Metabolites with lower abundance in FLT3-ITD are in italics.
Similar to analysis in plasma samples, multivariate analyses were conducted to evaluate the effectiveness of discriminating cellular metabolome by FLT3 status. PCA did not differentiate FLT3 groups (Fig. 2A), but PLSDA analysis showed clear discrimination between FLT3-WT and FLT3-ITD groups (Fig. 2B). The Q2 score was 0.35 in cell metabolome model indicating clear classification of two groups. Heat-Map of 208 cellular metabolites with significantly differential abundance between FLT3-ITD and FLT3-WT patients is shown in Fig. 2C and D shows all the metabolites detected in cell samples.
We acknowledge the fact that genetic and cytogenetic lesions within these patients, specifically the higher frequency of normal cytogenetics within FLT3-ITD patients as compared to FLT3-WT, could partially contribute to the observed differences. Given the limited sample size, the sub-classification of FLT3-status by cytogenetics requires future studies in a bigger patient cohort. However, our analysis of patients with normal cytogenetics showed consistent results when analyzed by FLT3 status (63% of metabolites in plasma and 76% in cells overlapped with the whole date set, data not shown).
Pathway analysis of metabolites identified in the FLT3-ITD vs. FLT3-WT
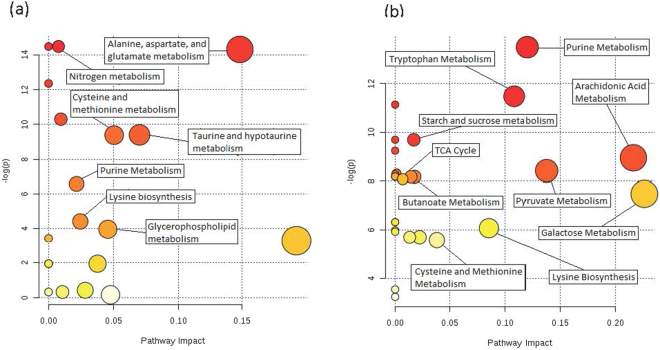
Pathway analysis was performed using MetaboAnalyst software. The pathway library included 80 known human metabolic pathways. Significance threshold was set at FDR <0.05. Pathway analysis in the plasma metabolome identified 10 significantly impacted metabolic pathways (Fig. 3a) and included pathways such as cysteine and methionine metabolism, purine metabolism and biosynthesis, and metabolic pathways of several amino acids. Pathway analysis of the cell metabolome identified 22 significantly impacted metabolic pathways (Fig. 3b). Of interest were metabolites involved in purine metabolism or biosynthesis pathways and pyruvate metabolism that were significantly influenced by the FLT3-status. Pathways of interest associated with disease progression in both patient samples forms include purine and cysteine and methionine metabolism.
Figure 3.
(a) Pathway analysis of significantly differential metabolites in plasma samples. Several of the most significantly impacted metabolic pathways have been labeled. Several impacted pathways reflected changes in the abundance of amino acids and nucleotides. (b) Pathway analysis of significantly differential metabolites in cell samples. The cell metabolome showed enrichment of changes to organic acid metabolic pathways. Color of circles indicate levels of statistical significance, with darker red reflecting smaller p-values and lighter colors down to white reflecting larger p-values. Circle size is meant to reflect pathway impact values as determined by pathway topology analysis. Larger circles indicate a more highly impacted metabolic pathway and smaller circles indicated a less impacted pathway.
We evaluated the metabolites using KEGG database and overall the purine pathway (http://www.genome.jp/kegg-bin/show_pathway?map = map00230&show_description = show) showed a total of six metabolites with significantly different abundance between FLT3-WT and FLT3-ITD patients in both plasma and cell samples indicating differences in purine analogs levels. Further cysteine and methionine metabolism pathway (http://www.genome.jp/kegg-bin/show_pathway?map = map00270&show_description = show) were impacted in both plasma and cell samples with three metabolites S-Sulfo-L-Cysteine (Cysteine-S-Sulfate) and L-Cysteate (L-Cysteic acid) in plasma samples and L-methionine in cell samples with differential abundance. The cysteine and methionine metabolic pathway is very closely linked to several pathways that were also significantly impacted by FLT3 status, including taurine and hypotaurine metabolism; glycine, serine and threonine metabolism; and sulfur metabolism. The citrate cycle (http://www.genome.jp/kegg-bin/show_pathway?map = map00020&show_description = show) was significantly impacted in cell samples through difference in succinate abundance.
Discussion
AML is a very heterogeneous disease with diverse clinical and biological features. One of the features associated with poor outcome is the presence of mutations, specifically an internal tandem duplication in FLT3, also known as FLT3-ITD. FLT3 has been implicated in stem cell differentiation and proliferation. Presence of FLT3-ITD with greater mutation burden (measured as higher ITD/WT allelic ration) is a well-established prognostic factor with greater allelic burden associated with significantly poor outcome. Gene expression studies to enhance understanding of FLT3 biology have identified patterns of expression differences associated with presence of FLT3-ITD. One of the newer and emerging approaches to enhance understanding of disease biology or identify biomarkers of response is metabolomics. In this study, we report the first metabolomics analysis of plasma and cell samples from pediatric AML patients with FLT3-ITD or FLT3-WT AML. Although overall FLT3-ITD status did not define the global leukemia metabolome, we identified multiple metabolic features with differential abundance between FLT3-ITD and FLT3-WT AML. While the contribution of other cytogenetic differences within FLT3-ITD and FLT3-WT cannot be ruled out, our results show multiple nucleosides and their intermediates, amino acids, organic acids, and lipids with role in cell proliferation and cancer progression to be differential in levels by FLT3 status.
Of significant interest was higher abundance of nucleosides such as guanine, hypoxanthine, inosine, adenosine, guanosine, and adenosine 5′-monophosphate within FLT3-ITD. Greater number of nucleosides within leukemic cells in FLT3-ITD as compared to FLT3-WT suggests propensity of greater cellular proliferation by providing nucleotides for DNA and RNA synthesis. Purine analogs such as ATP and GTP have been also shown to be crucial in providing cellular energy and intracellular signaling, with a role in cancer progression19.
Our results show that cellular metabolome of FLT3-ITD AML is less abundant in tryptophan and 5-formyl-hydroxykynurenamine. Tryptophan is converted to N-formyl kynurenine by Indolamine 2,3 dioxygenase (IDO) and to kynurenine through formamidase. IDO levels as well as kynurenine/tryptophan levels have been associated in several malignancies including AML20,21. Our results, although consistent with these previous findings, demonstrate the need for further in depth investigation of tryptophan metabolic pathway in context of FLT3-ITD AML. Among other amino acids, methionine was found to be more abundant in FLT3-ITD AML. L-methionine has been indicated in oncogene activation, as multiple studies have shown an increase in methionine metabolism in several cancer cell lines22 and patients23 to the extent that they are dependent on the amino acid as an energy source for cell growth. Succinate, a player in the citrate cycle, has been shown to inhibit 2-oxoglutarate dependent histone and DNA methylate enzymes, thus impacting epigenetic regulation. Given that DNA methylation is one of the hallmarks of leukemogenesis, metabolomic rewiring due to differential abundance of metabolites such as succinate or the well-established oncometabolite 2-hydroxyglutarate etc. can potentially contribute to leukemogenesis. This opens up opportunities to investigate relationships between differential succinate levels with epigenetic deregulation24.
L-carnitine was observed to be significantly higher in FLT3-ITD. It is involved in the transport of fatty acids across the mitochondrial membrane with the assistance of carnitine palmitoyl transferase thus allowing for cellular energy production through fatty acid β-oxidation. Serum profiling had previously shown a similar increase in carnitine abundance for patients with non-small-cell lung cancer as compared to healthy controls25. In addition, a recent metabolomics study reported increased carnitine levels with poor metastasis free survival outcomes in patients with different forms of soft tissue sarcoma26. These data suggests carnitine as a common thread among several cancer metabolomics studies, and our results further indicate that carnitine might be a useful target in future biomarker studies for FLT3-ITD pediatric AML.
Our results show lysophopholipids with differential abundance in FLT3-ITD cell samples. Lysophospholipids have been associated with disease progression in several forms of cancer, including AML, by upregulating signaling pathways27,28. Autotaxin, a lysophospholipid converting enzyme, in particular has been shown to have an increased activity that is associated with increased cell proliferation and migration in FLT3-ITD AML29 and might be potentially contributing to differential levels of LysoPEs. Overall, our results suggest differences in amino acid, purine metabolic pathways fatty acid metabolism/synthesis and various intermediates of glycolysis and TCA cycle to be enriched in FLT3-ITD vs. FLT3-WT AML.
To the best of our knowledge this report presents the first metabolomic evaluation of FLT3-ITD and FLT3-WT in pediatric AML. Our results demonstrate differential abundance of metabolites mapping to multiple pathways of biological relevance between FLT3-ITD patients as compared to FLT3-WT, these results were true for both diagnostic plasma and leukemic cell specimens. Although the global leukemia metabolome is not differentiated by FLT3 status, our results establish a precedent that certain metabolic features may differ by disease risk group categories and may have potential to be utilized as biomarkers for risk group classification. We recognize that the sample size of 16 patients is relatively small and may not adequately represent the pediatric AML patient population thus warranting a need for larger studies in pediatric AML to enhance our understanding of the biology underlying different risk group features and differential outcome. Our results open up opportunities to further enhance our understanding of FLT3 biology and ultimately identify novel drug targets that might be used to improve treatment outcome in AML.
Materials and Methods
Study Population
Plasma and cell samples were obtained from specimens obtained at diagnosis prior to initiation of any chemotherapy from 16 patients (8 with FLT3-ITD and 8 with FLT3-WT) enrolled in the Children’s Oncology Group-AAML1031 Clinical Trial (NCT01371981). The mononuclear cells were isolated using Ficoll-Hypaque density-gradient centrifugation. The blast percentage was >60% at the time of diagnosis and specimen collection. FLT3-ITD mutational analysis is routinely performed by Children’s Oncology Group reference labs using methods described previously30–32. Briefly, for mutational analysis exons 11 and 12 of FLT3 were amplified by PCR using primers 12 R, 5′-CTTTCAGCATTTTGACGGCAACC-3′, and 11 F, 5′-GCAATTTAGGTATGAAAGCCAGC-3′ followed by sequencing of bands resolved using 5% polyacrylamide gel32. All the patient samples with aberrant FLT3 amplification were confirmed by sequencing before post-mutational analysis for FLT3-ITD allelic ratio. FLT3-ITD +ve samples were further examined using Genescan analysis (Applied Biosystems, Foster City, CA) to determine the ratio between mutant and WT-FLT3 using previously described method31. Allelic ratio was calculated by dividing the peak height of the ITD product to that of the normal WT product. In cases in which more than one ITD product was present, ITD peak heights were added and divided by the WT peak height. The allelic ratio for FLT3-ITD patients was >0.80 (median allelic ratio: 1.11). AAML1031 is a randomized Phase III clinical trial that recruited newly diagnosed pediatric AML patients, aged 30 years and younger and is conducted in accordance with the Declaration of Helsinki and is registered as NCT01371981 at www.clinicaltrials.gov. The study is approved by the institutional review board (IRB) at Fred Hutchinson Cancer Center and Children’s Oncology Group-Myeloid committee and informed consent or assent has been obtained from patients or parents as appropriate. Patient characteristics are summarized in Table 1, detailed information is provided in Supplementary Table 3.
Sample Preparation
Plasma samples were aliquoted to 100 µL volumes. 20 µL of Internal Standard Solution (Creatine-D3, L-Leucine-D10, L-Tryptophan-D3, Caffeine-D3, Leucine 13C6, L-Tyrosine 13C6, Phenylalanine 13C6, L-Tryptophan-2,3,3-D3, Aspartic Acid_t-boc, Leucine_t-boc, Propionic acid 13C3, Succinic acid_d4, Tyrosine 13C6, and Salicylic Acid-D4) and 800 µL of precipitate solution (8:1:1 acetonitrile:methanol:acetone) were added to each sample to attain a 1:8 sample to solvent ratio. Samples were vortexed and placed in 4 °C refrigerator for 30 min to precipitate proteins. Proteins were isolated and removed through centrifugation at 20,000 g for 10 min at <10 °C. 750 µL of clear supernatant were collected and placed in individual collection tubes. Supernatants were dried using nitrogen gas (N2 Dryer, Organomation Associates Inc., Berlin, MA). Isolated metabolites were reconstituted with addition of 100 µL of reconstitution solution (Phenylalanine_t-boc, Tryptophan_t-boc, Tyrosine_t-boc in 0.1% formic acid in water). Reconstituted metabolites were chilled on ice bath for 15 min and centrifuged using previous settings. Supernatants were collected and placed in glass liquid chromatography vials for autosampling.
Cell samples were isolated to 1 × 106/mL pellets for preparation. Cell pellets were washed using 1 mL of 40 mM ammonium formate in water solution. Samples were vortexed and centrifuged at 2000 rpm for 5 min at 5 °C. Supernatant was discarded. The washing procedure was repeated twice for a total of three washes. 50 µL of 5 mM ammonium acetate in water solution was added to the cell pellets. Cell samples were then homogenized using Bead Beater (Fastprep 96, MPBio, Santa Ana, CA) at 1800 rpm for 30 sec. Samples were incubated at 4 °C for 30 min. 2 µL of internal standard solution and 1 mL of 80% methanol in water solution were added to each sample. Samples were homogenized once more using Bead Beater set to 1800 rpm for 30 sec and incubated at 4 °C for 10 min. Samples were then centrifuged at 2000 rpm for 5 min at 5 °C. 1000 µL of supernatant were collected and placed in individual collection tubes. Supernatants were dried using nitrogen gas (N2 Dryer, Organomation Associates Inc.). Isolated metabolites were reconstituted with addition of 30 µL of reconstitution solution. Reconstituted metabolites were chilled on ice bath for 15 min and centrifuged using previous settings. Supernatants were collected and placed in glass liquid chromatography vials for autosampling.
Metabolomics Analysis
Global metabolomics profiling was performed on a Thermo Q-Exactive Oribtrap mass spectrometer with Thermo Ultimate 3000 UHPLC (Thermo, San Jose, CA) at Southeast Center for Integrated Metabolomics (SECIM) University of Florida. All samples were analyzed in positive and negative heated electrospray ionization with a mass resolution of 35,000 at m/z 200 as separate injections. Separation was achieved on an ACE 18-pfp 100 × 2.1 mm, 2 µm column with mobile phase A as 0.1% formic acid in water and mobile phase B as acetonitrile. This is a polar embedded stationary phase that provides comprehensive coverage, but does have some limitation is the coverage of very polar species. The flow rate was 350 µL/min with a column temperature of 25 °C. 4 µL was injected for negative ions and 2 µL for positive ions.
Data Processing and Statistical Analysis
Raw data files were converted to mzxml file format using MS Convert software (ProteoWizaed 3.0). MZmine 2 was used to identify features, deisotope, align features and perform gap filling to fill in any features that may have been missed in the first alignment algorithm. All adducts and complexes were identified and removed from the data set. The data was searched against SECIM’s internal retention time metabolite library of 1100 compounds. Positive and negative mode data sets including known and unknown metabolites were merged and subjected to statistical analyses. Peak intensity values for each patient sample were grouped according to patient’s recorded FLT3 status (WT or ITD) for categorical analysis. Metabolomics statistical analysis was performed on MetaboAnalyst 3.0 web based software17. Datasets used for analysis were composed of merged peak area data generated by both positive and negative ionization. The metabolomics datasets included all known and unknown features detected through UHPLC-MS for analysis. Prior to analysis, MetaboAnalyst was used for data filtration and normalization functions. Missing value estimation was performed with metabolites missing over 30% of values removed. Remaining missing values were replaced by a calculated small value (half of the minimum positive value in the original data). Data filtering was performed using interquartile range. Data processing included normalization by sum, log transformation, and pareto scaling. Processed metabolomics data were then analyzed using univariate, multivariate, and clustering methods. Univariate analyses included t-test and fold change analysis. False discovery rate (FDR) was calculated to account for multiple hypothesis correction. Significance threshold was set at FDR <0.05. Multivariate analyses included principal component analysis (PCA) and partial least square discriminatory analysis (PLSDA). Clustering analysis included generation of heatmaps.
Metabolic Pathway Enrichment and Topology Analysis
Peak intensity values for metabolites with significantly different abundance in plasma and cell samples were imported into MetaboAnalyst pathway analysis function, which generated integrated pathway enrichment and pathway topology analyses. Metabolite identifiers were converted as necessary according to synonyms listed in the human metabolome database (HMDB). The pathway impact measurement represented the sum of importance measures, generated by topology analysis, of significant metabolites normalized by importance measures of all metabolites in the associated pathway. Additionally we utilized KEGG database to map the significant metabolites on the pathways of interest33–35. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank University of Florida SECIM for providing resources to perform metabolic profiling and Children’s Oncology Group for supporting the study. This research was supported by NIH under the award numbers: U10CA180899, U10CA180886, U10CA98413, and U10CA098543 as well as by NCI-R01CA132946.
Author Contributions
B.S. and J.K.L. designed the study and B.S., T.G., J. G.-C. and J.K.L. were involved in data analysis and processing. J.-G.C. performed LC-MS studies S.M. provided clinical expertise. All the authors contributed to data analysis, interpretation and manuscript writing.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23863-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maroc N, et al. Biochemical characterization and analysis of the transforming potential of the FLT3/FLK2 receptor tyrosine kinase. Oncogene. 1993;8:909–18. [PubMed] [Google Scholar]
- 2.Saultz, J. N. & Garzon, R. Acute Myeloid Leukemia: A Concise Review. J Clin Med5 (2016). [DOI] [PMC free article] [PubMed]
- 3.Bullinger L, Dohner K, Dohner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J Clin Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 4.Medinger M, Lengerke C, Passweg J. Novel Prognostic and Therapeutic Mutations in Acute Myeloid Leukemia. Cancer Genomics Proteomics. 2016;13:317–29. [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15:e382–94. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 6.Dohner H, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 7.Medinger M, Lengerke C, Passweg J. Novel therapeutic options in Acute Myeloid Leukemia. Leuk Res Rep. 2016;6:39–49. doi: 10.1016/j.lrr.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medinger, M. & Passweg, J. R. Acute myeloid leukaemia genomics. Br J Haematol (2017). [DOI] [PubMed]
- 9.Smith CC, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–3. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, et al. Serum metabolomic profiling of prostate cancer risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Br J Cancer. 2016;115:1087–1095. doi: 10.1038/bjc.2016.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budczies J, et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism. J Proteomics. 2013;94:279–88. doi: 10.1016/j.jprot.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 12.MacIntyre DA, et al. Serum metabolome analysis by 1H-NMR reveals differences between chronic lymphocytic leukaemia molecular subgroups. Leukemia. 2010;24:788–97. doi: 10.1038/leu.2009.295. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Zhang H, Sun X, Sun C, Ren L. Biomarker identification and pathway analysis by serum metabolomics of childhood acute lymphoblastic leukemia. Clin Chim Acta. 2014;436:207–16. doi: 10.1016/j.cca.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Tiziani S, et al. Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS One. 2009;4:e4251. doi: 10.1371/journal.pone.0004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen WL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–54. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JH, et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci USA. 2013;110:17017–22. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa T, Ohki T, Maeda K, Saito A, Kobayashi K. Pattern of aliphatic dicarboxylic acids in uremic serum including a new organic acid, 2,4-dimethyladipic acid. Clin Chim Acta. 1979;99:71–83. doi: 10.1016/0009-8981(79)90141-4. [DOI] [PubMed] [Google Scholar]
- 19.Pedley AM, Benkovic SJ. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curti A, et al. Acute myeloid leukemia cells constitutively express the immunoregulatory enzyme indoleamine 2,3-dioxygenase. Leukemia. 2007;21:353–5. doi: 10.1038/sj.leu.2404485. [DOI] [PubMed] [Google Scholar]
- 21.Hara T, et al. Prognostic value of the combination of serum l-kynurenine level and indoleamine 2,3-dioxygenase mRNA expression in acute myeloid leukemia. Leuk Lymphoma. 2016;57:2208–11. doi: 10.3109/10428194.2015.1128541. [DOI] [PubMed] [Google Scholar]
- 22.Cellarier E, et al. Methionine dependency and cancer treatment. Cancer Treat Rev. 2003;29:489–99. doi: 10.1016/S0305-7372(03)00118-X. [DOI] [PubMed] [Google Scholar]
- 23.Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–36. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Pollard PJ. Succinate: a new epigenetic hacker. Cancer Cell. 2013;23:709–11. doi: 10.1016/j.ccr.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Klupczynska A, et al. Study of early stage non-small-cell lung cancer using Orbitrap-based global serum metabolomics. J Cancer Res Clin Oncol. 2017;143:649–659. doi: 10.1007/s00432-017-2347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou S, Balluff B, Cleven AH, Bovee JV, McDonnell LA. Prognostic Metabolite Biomarkers for Soft Tissue Sarcomas Discovered by Mass Spectrometry Imaging. J Am Soc Mass Spectrom. 2017;28:376–383. doi: 10.1007/s13361-016-1544-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SC, et al. Autotaxin and LPA1 and LPA5 receptors exert disparate functions in tumor cells versus the host tissue microenvironment in melanoma invasion and metastasis. Mol Cancer Res. 2015;13:174–85. doi: 10.1158/1541-7786.MCR-14-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SJ, et al. Lysophosphatidylethanolamine utilizes LPA(1) and CD97 in MDA-MB-231 breast cancer cells. Cell Signal. 2013;25:2147–54. doi: 10.1016/j.cellsig.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Ortlepp C, et al. Autotaxin is expressed in FLT3-ITD positive acute myeloid leukemia and hematopoietic stem cells and promotes cell migration and proliferation. Exp Hematol. 2013;41:444–461 e4. doi: 10.1016/j.exphem.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Meshinchi S, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–61. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwaan CM, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood. 2003;102:2387–94. doi: 10.1182/blood-2002-12-3627. [DOI] [PubMed] [Google Scholar]
- 32.Meshinchi S, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97:89–94. doi: 10.1182/blood.V97.1.89. [DOI] [PubMed] [Google Scholar]
- 33.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.