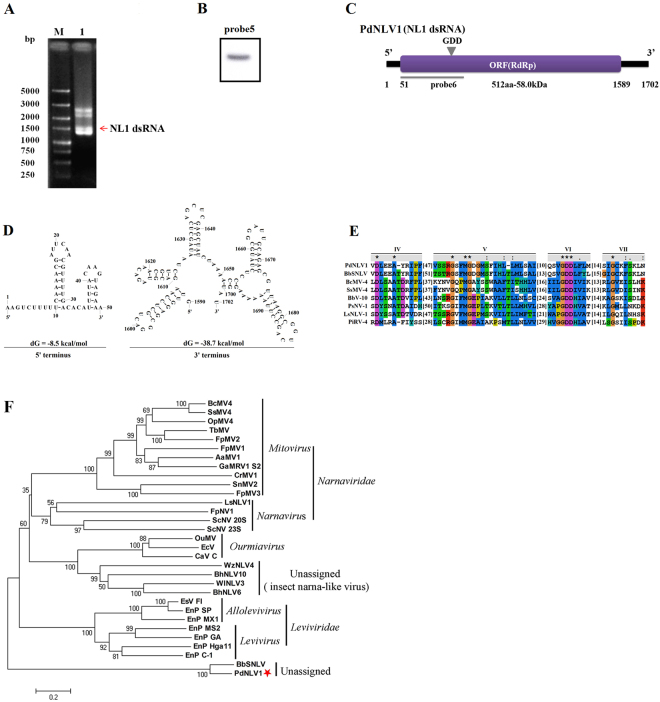
Figure 2.
Molecular characterization and evolutionary analysis of PdNLV1. (A) Electrophoretic profile on a 1% agarose gel of dsRNA preparations extracted from P. digitatum isolate HS-RH2 after digestion with DNase I and S1 nuclease, and stained with ethidium bromide (lane 1). The dsRNA of PdNLV1 was indicated by red arrows. Lane M, DNA size marker (DS 5000, TaKaRa, Dalian, China). (B) Northern blot analysis of PdNLV1 dsRNA extracted from P. digitatum isolate HS-RH2 (Fig. S7). The position of the specific probe (probe 5) was shown in panel C. (C) Schematic representation of the genetic organization of PdNLV1. (D) Predicted secondary structure of the PdNLV1 5′- and 3′-UTRs with the lowest energy. (E) Comparison of the conserved motifs of RdRps encoded by PdNLV1 and other selected mycoviruses. Four conserved RdRp motifs corresponding to motifs IV, V, VI and VII were shown. The asterisks signified identical amino acid residues, colons signified highly conserved residues, and single dots signified less conserved but related residues. Numbers within the brackets indicated the number of aa not shown. (F) Phylogenetic analysis of PdNLV1 RdRp and other selected RdRps. Members of narnaviruses and Narna-like viruses in databases (Table S2) were selected, and the phylogenetic trees for RdRp sequences were constructed using the neighbor-joining method of MEGA version 6 software with bootstrapping analysis of 1000 replicates.