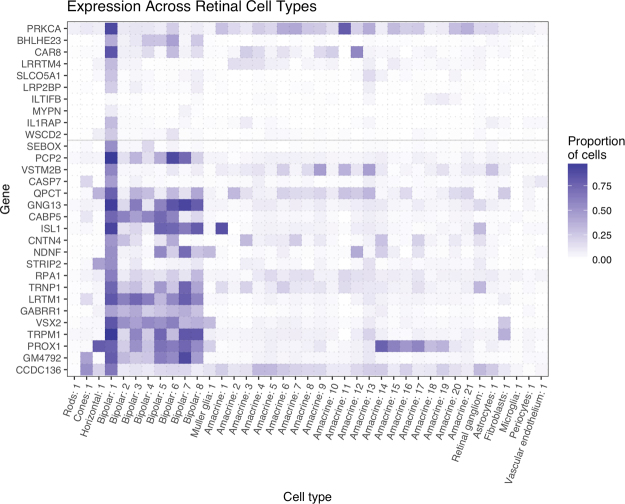
Figure 6.
Relative expression of genes identified in the adult rod bipolar cell transcriptome in other retinal cell types. Heat map showing relative expression value (normalized by row) for genes, identified in the adult RB transcriptome, in other retinal cell types using published Drop-seq data for the whole retina17. The upper section shows RB genes identified in the Bhlhe23−/− mouse model; the lower section includes the 22 genes found in all 4 adult RBC transcriptomes (Fig. 4). Intensity of colour shows the total proportion of cells, for that cell type, which express the gene of interest. ‘Bipolar 1′ was inferred to represent RBCs due to the high expression of Prkca. The results show that Car8, Lrp2bp, Slco5a1, Lrrtm4, and Mypn were all strongly expressed in the cluster identified as RBCs, although none were exclusive to RBCs. Bhlhe23 is an established marker of RBCs but was not specifically expressed by RBCs in the Drop-seq profiling data of different retinal cell types17. In addition, Iltifb was expressed only at low levels in RBCs identified in the Drop-seq profiling data of different retinal cell types17, but occurred in the rod bipolar transcriptome we identified in our study and the Drop-seq profiling data of different bipolar subtypes18 (see Fig. 4b). Our results for IL1RAP and WSCD2 immunolabelling (Fig. 5i–k) suggest that these transcripts are either expressed at a low level in RBCs, not transcribed and/or the RBC data from Drop-seq profiling of different retinal cell types published by Macosko et al.17 was contaminated by a small number of ACs.