Fig. 7.
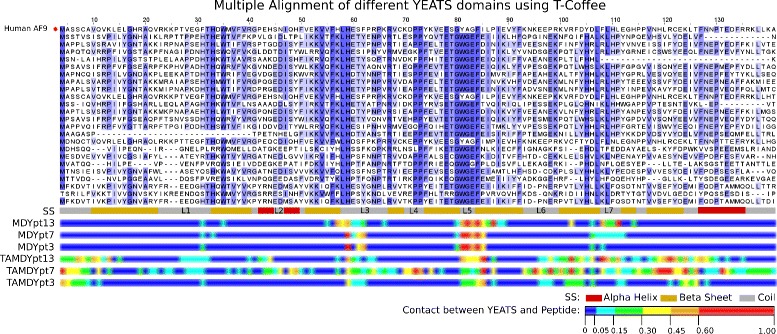
Sequence alignments. Multiple alignment between the human AF9 YEATS sequence (Uniprot ID: P42568) and other YEATS sequences (Uniprot IDs, from top to bottom: Q59LC9, Q6FXM4, Q4I7S1, Q4PFI5, Q4WPM8, Q10319, Q755P0, Q6CIV8, A2AM29, P0CM08, P53930, Q5BC71, Q7RZK7, Q6CF24, Q03111, Q99314, F4IPK2, P35189, Q9FH40, O94436, Q9CR11, Q9ULM3, O95619), colored from white to blue according to the conservation of the residue. The alignment was done using T-Coffee [54]. The columns corresponding to gaps in the human AF9 sequence were removed in order to reduce the figure width. Under the alignment, the first horizontal bar represents the secondary structure of the AF9 YEATS NMR structure (PDB 2NDF: α helices in red and β strands in orange). The following six bars represent the proportion of contact of each YEATS residue with the peptide, from blue (near 0% of maximum number of contacts) to red (over 80% maximum number of contacts). There is one bar for each trajectory: MDYpt13, MDYpt7, MDYpt3, TAMDYpt13, TAMDYpt7, TAMDYpt3. Contacts were computed with a python script using the MDAnalysis module [52]
