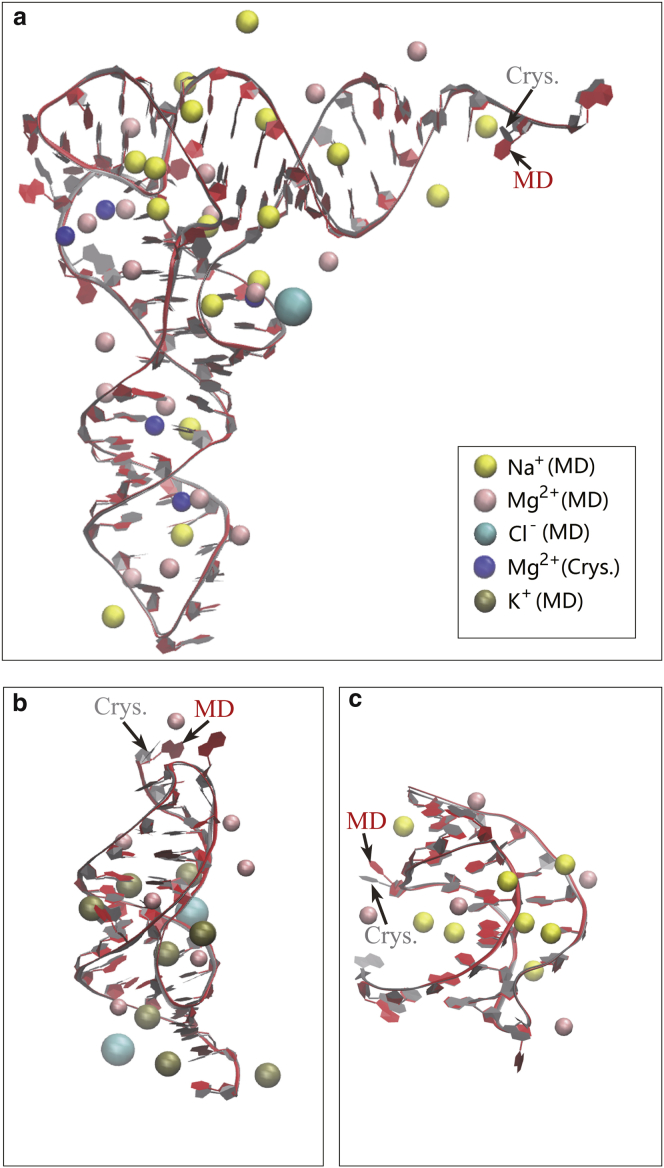
Figure 1.
Comparison between the PDB crystal structure and MD snapshot structures of the three RNA models: (a) the yeast phenylalanine tRNA (PDB: 1TRA); (b) a gene 32 messenger RNA pseudoknot of bacteriophage T2 (PDB: 2TPK); and (c) BWYV, a ribosomal frameshifting viral pseudoknot (PDB: 437D). The RNA contour structures are drawn using NewRibbons of VMD (Theoretical and Computational Biophysics Group) (83). The crystal structure (Crys.) and the MD simulated structure (MD) are very close except for the orientations of a few bases. The figure shows the simulated distributions of the different metal ions: Na+, K+, Mg2+ (MD), and Cl−. Also shown in (a) are the five Mg2+ ions (Crys.) observed in the crystal structures. To see this figure in color, go online.