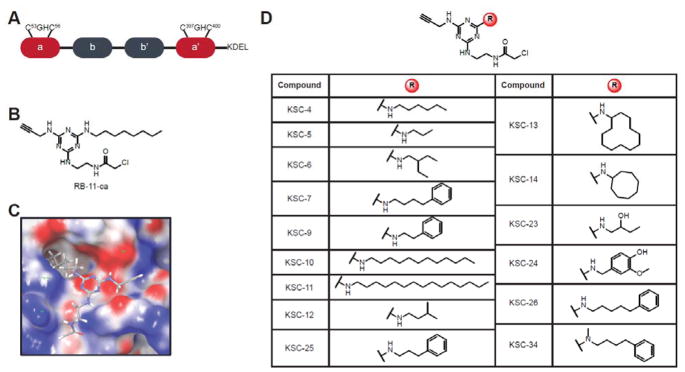
Figure 1.
Domain organization of PDIA1 and chemical structures of PDIA1 inhibitors. (A) PDIA1 comprises two active site a-type domains that contain a CGHC active site motif, together with two b-domains implicated in substrate recognition and binding. (B) Chemical structure of RB-11-ca, a previously reported a-site inhibitor of PDIA1. RB-11-ca contains a central triazine scaffold, a chloroacetamide reactive group for covalent cysteine modification, an alkyne bioorthogonal handle for CuAAC, and an octylamine diversity element. (C) Predicted binding pose of RB-11-ca in a domain active site of PDIA1 by the covalent docking algorithm from Schrodinger, Inc. (D) Structures of second-generation PDIA1 inhibitors obtained by varying the diversity element of RB-11-ca.