Fig. 2. Functional heterogeneity of MPP lineage fates in steady state hematopoiesis.
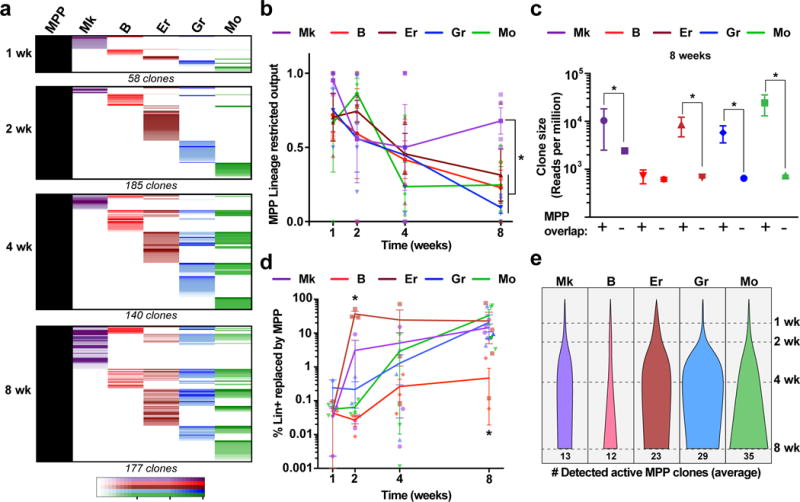
a, Chart shows the alignment of all active MPP tags together with the five analysed blood lineages at each time point (all tags collected from 3 mice per time point). LT-HSC tags were analysed in parallel and excluded from the analysis to represent only MPP behaviour. b, Fraction of active MPP tags that overlap with a single lineage (calculated independently for each lineage). Values are mean +/− s.e.m. from 3 mice. *pMk-Er=0.13, pMk-Gr=0.03, pMk-Mo=0.03, pMk-B=0.001 (8 wk). c, Distribution of Lin+ clone sizes comparing tags overlapping with MPP vs. non-overlapping at 8 wk. Values are median and interquartile ranges of all detected clones from 3 mice. *Kolmogorov-Smirnov pMk=0.03, pB=0.25, pEr=0.03, pGr=0.001, pMo=0.003. d, Fraction of each lineage replaced by MPPs calculated as the percentage of total MPP-overlapping lineage reads over time. Values are mean +/− s.e.m. from 3 independent mice. *pEr-Gr/Mo/B=0.04, pEr-Mk=0.03 (2 weeks) and pB-Er/Mk=0.03, pB-My=0.04 (8 weeks). e, Average number of detected active MPP clones per lineage per mouse at different time points (normalized for %DsRed labelling efficiency).
