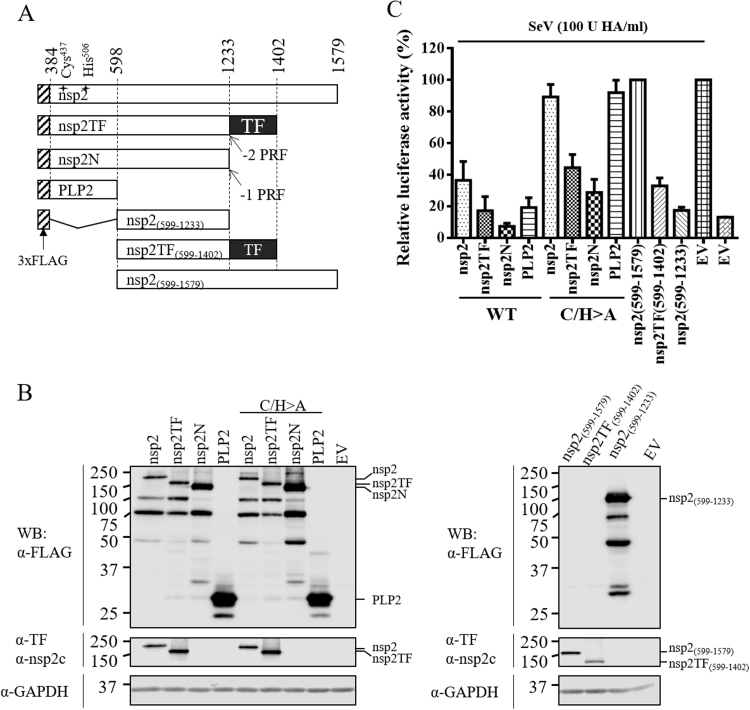
Fig. 1.
PRRSV nsp2 variants suppress type I IFN production. (A) A schematic diagram of individual PRRSV nsp2 variants (nsp2, nsp2TF and nsp2N) and domains [PLP2, nsp2(599–1233), nsp2TF(599–1402) and nsp2(599–1579)]. The numbers show both N- and C-terminal residues on polyprotein 1a for each individual protein. The catalytic residues of the PLP2 domain are labeled as Cys437 and His506. (B) Expression of nsp2-related proteins were evaluated by western blot analysis using anti-FLAG M2 mAb. GAPDH was detected as loading control. Rabbit pAb against the unique nsp2 C-terminus was used to confirm the expression of nsp2 and nsp2(599–1579), while rabbit pAb against unique nsp2TF C-terminus was used to confirm the expression of nsp2TF and nsp2TF(599–1402). (C) Effect of nsp2-related proteins on the expression of IFN-β promoter-driven luciferase expression. HEK-293T cells were cotransfected with a plasmid expressing individual nsp2-related proteins or domains, p125-Luc reporter plasmid expressing firefly luciferase under the control of the IFN-β promoter (0.5 μg), and pRL-SV40 reporter plasmid (20 ng). An empty vector (EV) was used as a control. At 24 h posttransfection, cells were stimulated with SeV at 100 HA units/mL or for 16 h. Cell lysates were harvested for measuring luciferase activity. Relative luciferase activities were calculated by normalizing the firefly luciferase to Renilla luciferase activities; the relative luciferase activity in cells with EV-transfection and SeV stimulation was set as 100%. The mean value and SEM of representative experiments are shown. All experiments were repeated at least three times, and duplicates were performed each time.